Amplification of plant disease-resistance genes in pepper is intimately linked to transposon activity
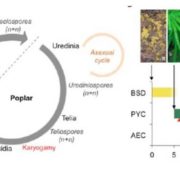
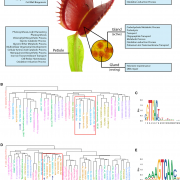
 Peppers are an economically and ecologically important crop plants, but genomic resources are rather scarce. The authors provide here new reference genome sequences for three species of hot pepper (Capsicum baccatum, C. chinense and C. annuum), identifying evolutionary forces that have shaped pepper genomes. Based on these new resources, the study reveals important species-specific chromosomal rearrangements, but also massive amplification of retrotransposons and specific gene families including the nucleotide-binding and leucine-rich-repeats proteins (NLRs), the major disease-resistance gene families in plants. By comparing proliferation dynamics of NLRs in pepper and other plant genomes, the authors show that a large proportion of the NLRs have been created through retroduplication, a process involving the reverse transcription of the NLRs from their mRNA into a complementary DNA copy, mediated by enzymes encoded by retrotransposons. Altogether, this study stresses out the intimate link between genes and transposons in the evolution of genomes, as well as the necessity of high-quality genome references for in-depth genomic studies. (Summary by Matthias Benoit) Genome Biol. 10.1186/s13059-017-1341-9
Peppers are an economically and ecologically important crop plants, but genomic resources are rather scarce. The authors provide here new reference genome sequences for three species of hot pepper (Capsicum baccatum, C. chinense and C. annuum), identifying evolutionary forces that have shaped pepper genomes. Based on these new resources, the study reveals important species-specific chromosomal rearrangements, but also massive amplification of retrotransposons and specific gene families including the nucleotide-binding and leucine-rich-repeats proteins (NLRs), the major disease-resistance gene families in plants. By comparing proliferation dynamics of NLRs in pepper and other plant genomes, the authors show that a large proportion of the NLRs have been created through retroduplication, a process involving the reverse transcription of the NLRs from their mRNA into a complementary DNA copy, mediated by enzymes encoded by retrotransposons. Altogether, this study stresses out the intimate link between genes and transposons in the evolution of genomes, as well as the necessity of high-quality genome references for in-depth genomic studies. (Summary by Matthias Benoit) Genome Biol. 10.1186/s13059-017-1341-9