Alternative Splicing Plays a Major Role in Plant Response to Cold Temperatures
Plants in temperate regions experience near-freezing temperatures that allow them to develop a cold response prior to freezing. This cold acclimation process involves changes to chromatin structure, transcription, RNA processing, translation, post-translational modifications and protein stability. Genome-wide studies of cold acclimation have largely focused on changes to transcription. Several pathways are known to undergo changes in transcriptional regulation following cold exposure including the Cold-Repeat Binding Factor (CBF) signaling pathway and circadian cycles. Alternative splicing (AS) has also been implicated in cold acclimation, but the global impact of AS on cold responsive transcripts was previously unexplored (reviewed by Knight and Knight, 2012).
AS affects gene expression in a number of ways. Introns are removed from the majority of RNA transcripts by the spliceosome in consistent patterns. Under AS, those patterns change. AS can cause an RNA transcript to become unstable. For instance, the alternatively spliced transcript may contain a premature stop codon that triggers nonsense-mediated decay. Alternatively, the RNA might be translated into a protein with a slightly different function (for a description of the Arabidopsis AS landscape, see Marquez et al., 2012).
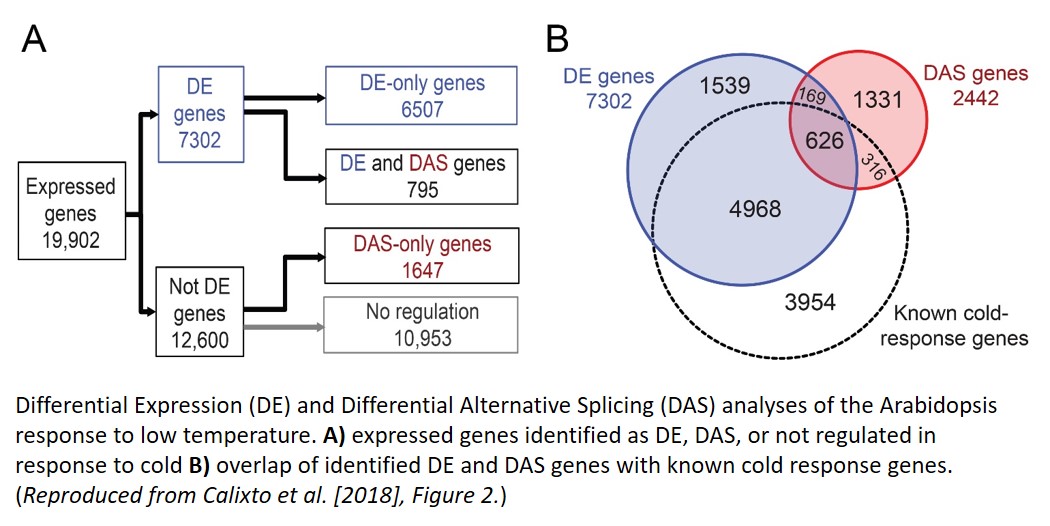
To analyze AS changes in response to cold while accounting for circadian effects, Calixto et al. (2018) developed an expanded reference transcriptome database for Arabidopsis, and then used time-series RNA-seq in conjunction with lowering temperatures. Using this global approach, they identified nearly 9,000 genes that were differentially regulated in response to cold, 3039 of which had not been recognized previously (see Figure). Differential expression of known cold response transcription factors, such as CBF genes, largely agreed with previous studies. 27% of the cold-responsive transcripts identified were alternatively spliced, with nearly two-thirds of these regulated only by AS (no differential expression at the gene level). The majority of differentially alternatively spliced (DAS) transcripts still led to a protein product.
 Differentially expressed genes that did and did not undergo AS clustered according to gene function. Differentially expressed genes that were not alternatively spliced were associated with general stress response, ribosome production, circadian cycling, photosynthesis, and lipid biosynthesis. DAS genes tended to be associated with splicing, chromatin binding, transcriptional regulation, signal transduction, and effects on the plasma membrane. AS impacted one third of the cold-responsive genes encoding transcription factors and nearly one half of the genes encoding splicing factors and other RNA-binding proteins. Many of these were known to be involved in stress response or circadian regulation, but previously had not been linked to cold response specifically.
Differentially expressed genes that did and did not undergo AS clustered according to gene function. Differentially expressed genes that were not alternatively spliced were associated with general stress response, ribosome production, circadian cycling, photosynthesis, and lipid biosynthesis. DAS genes tended to be associated with splicing, chromatin binding, transcriptional regulation, signal transduction, and effects on the plasma membrane. AS impacted one third of the cold-responsive genes encoding transcription factors and nearly one half of the genes encoding splicing factors and other RNA-binding proteins. Many of these were known to be involved in stress response or circadian regulation, but previously had not been linked to cold response specifically.
The DAS response was dynamic over time with AS patterns emerging for different genes throughout the time course. For many genes, DAS occurred very early, suggesting AS involvement in a rapid initial response to cold. 40% of DAS genes differed between day one at 4°C and day four at 4°C. Upon testing even shorter time and smaller temperature intervals, the authors detected AS changes as quickly as 40 minutes and after temperature changes of only two degrees. Of the “early AS” genes, four were known to be essential for cold tolerance. To determine whether others might be essential, Calixto et al. examined a knockout of the gene U2B”-LIKE, an RNA-binding protein which exhibited an early AS response and was not otherwise differentially expressed. These u2b”-like mutants were less cold tolerant and exhibited different AS patterns in five other genes, including one that is a major player in the CBF pathway.
The authors conclude that AS plays a major role in cold response. They show that AS impacts several cold-responsive transcription factors and RNA-binding proteins that had not been identified previously, many of which were regulated only by AS. The early response of some DAS genes such as U2B”-LIKE suggest that AS kickstarts a cascade of downstream effects that are important for initial response to cold and sustained cold tolerance. Intriguingly, AS is likely important for regulating other stress response pathways in addition to cold tolerance. The advanced RNA-seq analysis system described by Calixto et al could be used to explore AS in a variety of different contexts.
REFERENCES
Calixto, C.P.G., Guo, W., James, A.B., Tzioutziou, N.A., Entizne, J.P.,1,4, Panter, P.E., Knight, H., Nimmo, H.G., Zhang, R., and Brown, J.W.S. (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell. https://doi.org/10.1105/tpc.18.00177.
Knight, M.R., and Knight, H. (2012). Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 195: 737-751
Marquez, Y., Brown, J.W., Simpson, C., Barta, A., and Kalyna, M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22: 1184-1195.