A tug-of-war in the viral replication factory: How a viral protein interferes with stress granules
Hoffmann et al. explore replication of Cauliflower mosaic virus and how virus replication intersects RNA granule biology.
https://doi.org/10.1093/plcell/koad101
Gesa Hoffmann and Anders Hafrén, Swedish University of Agricultural Sciences
Background: Regulating RNA abundance and its availability for translation is one of the main struggles between virus and host during plant virus infections. The pararetrovirus Cauliflower mosaic virus (CaMV) shields its nucleic acids, proteins and particles from its host antiviral pathways by establishing large, amorphous condensates, termed viral factories. These condensates consist mainly of one multifunctional viral protein, the P6 protein. We have previously found that host RNA decapping proteins, canonically associated with phase-separated RNA granules, localize within these viral factories and aid in the translation of viral RNA. The interplay between virus replication and RNA granule biology is conserved among eukaryotes and likely represents an ancient mechanism.
Question: Our first aim was to further characterize the viral factory of CaMV and observe the behavior of host proteins within these condensates. Our second aim was to dissect the interaction of the viral P6 protein with RNA granules and RNA granule proteins, as well as its effect on translation in the context of condensation.
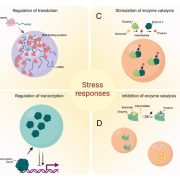
Findings: Unlike the rigid P6 matrix, RNA granule proteins can rapidly shuffle between viral factories and the surrounding cytoplasm. Several RNA granule proteins can bind viral RNA; however, their binding capacities seem to be specific for certain viral RNA species. Stress granule proteins preferentially bind to the non-coding highly abundant 8s RNA, which may dampen stress granule responses in the plant. The P6 protein, by contrast, strongly localizes to stress granules and hinders their establishment due to its remarkable ability to enhance global translation levels. Importantly, the efficiency with which P6 exerts its functions is in part coupled to its condensation level, which likely represents a self-attenuation mechanism of CaMV during prolonged infections.
Next steps: We have merely scratched the surface of the viral factory-localized host proteome and RNAome. Are there common features of the plant proteins and RNAs that accumulate within the viral factories? Elucidating which host factors are co-opted by the virus will further our understanding of virus disease in plants.
Reference:
Gesa Hoffmann, Silvia López-González, Amir Mahboubi, Johannes Hansonc, and Anders Hafrén (2023) Cauliflower mosaic virus protein P6 is a multivalent node for RNA granule proteins and interferes with stress granule responses during plant infection. https://doi.org/10.1093/plcell/koad101