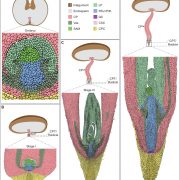
A single-cell Arabidopsis root atlas reveals developmental trajectories in WT and cell identity mutants (Dev. Cell)
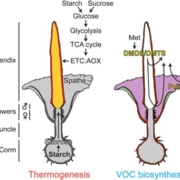
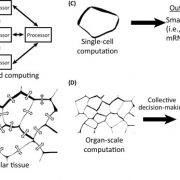
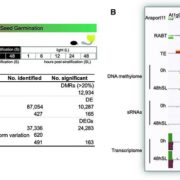
 Single-cell RNA sequencing (scRNA-seq) is a powerful tool to identify rare and novel cell types, characterize multiple cell types, and more accurately understand their functions across their life cycles. The Arabidopsis root is a perfect system in which to investigate cellular differentiation due to its immobile and organized cells. Shahan and Hsu et al. built a root scRNA-seq atlas (>110 000 cells) using an optimal transport-base method, StationaryOT. This method allows the reconstruction of developmental trajectories and helps to understand how the fate acquisition of a specific cell type is related to other cell types. The authors tested the utility of this method by performing scRNA-seq of shortroot (shr) and scarecrow (scr) mutants to identify unknown developmental phenotypes. scRNA-seq and in vivo experiments of scr suggest that young cells are cortex-like while older cells changes their identity to endodermis-like. These findings show how cell identity acquires attributes of other cell types over time. This work provides an outlook to test unknown cell-type specific markers and identify cell-type-specific responses to different environmental conditions. (Summary by Andrea Gómez Felipe @andreagomezfe) Dev. Cell 10.1016/j.devcel.2022.01.008
Single-cell RNA sequencing (scRNA-seq) is a powerful tool to identify rare and novel cell types, characterize multiple cell types, and more accurately understand their functions across their life cycles. The Arabidopsis root is a perfect system in which to investigate cellular differentiation due to its immobile and organized cells. Shahan and Hsu et al. built a root scRNA-seq atlas (>110 000 cells) using an optimal transport-base method, StationaryOT. This method allows the reconstruction of developmental trajectories and helps to understand how the fate acquisition of a specific cell type is related to other cell types. The authors tested the utility of this method by performing scRNA-seq of shortroot (shr) and scarecrow (scr) mutants to identify unknown developmental phenotypes. scRNA-seq and in vivo experiments of scr suggest that young cells are cortex-like while older cells changes their identity to endodermis-like. These findings show how cell identity acquires attributes of other cell types over time. This work provides an outlook to test unknown cell-type specific markers and identify cell-type-specific responses to different environmental conditions. (Summary by Andrea Gómez Felipe @andreagomezfe) Dev. Cell 10.1016/j.devcel.2022.01.008