WRKY1 integrates cellular nitrogen and light-energy resources in Arabidopsis thaliana
Tessa Moses
The Institute of Quantitative Biology, Biochemistry and Biotechnology (IQB3), The King’s Buildings, University of Edinburgh, Edinburgh EH9 3BF, United Kingdom.
E-mail: [email protected], ORCID ID: 0000-0001-9366-4727
WRKY1 is a member of the WRKY transcription factor family initially identified with roles in defense, but more recently shown to have diverse functions (Ülker and Somssich, 2004). The A. thaliana WRKY1 transcription factor has been implicated in nutrient signalling, where its expression has been reported to be repressed by organic nitrogen treatment (Gutiérrez et al., 2008) and activated by nitrogen starvation (Krapp et al., 2011). Furthermore, A. thaliana WRKY1 was among the genes regulated by light in a whole-transcriptome analysis of Arabidopsis that indicated that 35% of the genome is controlled by either light, nitrogen, or a combination of light and nitrogen signalling (Krouk et al., 2009).
In this issue of Plant Physiology, Heerah et al. (2019) report their investigation into the role of WRKY1 in coordinating responses to light and nitrogen signalling in A. thaliana through gene regulatory network analysis, genome-wide expression analysis, gene expression cluster analysis and metabolite pool analysis.
Using a minimum of one regulatory motif as a criterion for transcription factor binding in the promoter of putative target genes, a protein–DNA interaction network identified WRKY1 as directly associated with several nitrogen assimilation genes. Among these, GLUTAMATE DEHYDROGENASE (GDH1), NITRATE REDUCTASE 1 (NIA1) and NIA2 were predicted to be activated by WRKY1, while ASPARAGINE SYNTHETASE 1 (ASN1) was predicted to be repressed by WRKY1. ASN1 has also been reported to be regulated in response to light and carbon signalling (Thum et al., 2003). A genome-wide comparison of expression between wild type plants and three different wrky1 T-DNA mutant alleles causing 2–24% of wild-type WRKY1 expression identified 117 down-regulated genes involved in defense response and response to stress, and 256 up-regulated genes involved in response to carbohydrate and light stimuli, and regulation of nitrogen metabolism. In contrast to their expression in wild-type plants, genes for nitrate transporters and glutamate receptors were down-regulated in the wrky1 mutants, while ASN1 was up-regulated in the light period. The reprogramming of the nitrogen network and the presence of the W-box DNA elements that WRKY1 binds to in the promoters of these nitrogen metabolic genes support the regulatory role of WRKY1 in nitrogen signalling pathways (Heerah et al., 2019).
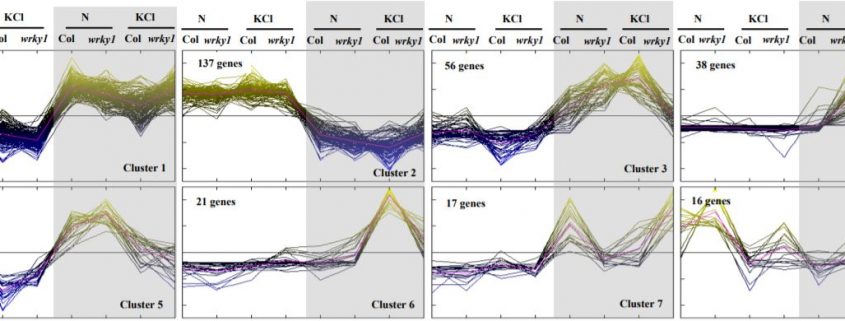
Gene expression cluster analysis of wild-type and wrky1 mutants grown in light or extended dark conditions identified the role of WRKY1 in regulating genes involved in light responses. In light conditions, WRKY1 repressed genes required to catabolise cellular resources and activated genes for the biosynthesis of energy-dependent metabolites, which contrasts with the activation of genes involved in respiration and repression of energy- expensive secondary metabolic processes by WRKY1 in the dark.
A similar nitrogen-dependent gene expression cluster analysis of wild-type and wrky1 mutants indicated that WRKY1 activates apoptosis genes and represses genes involved in protein biosynthesis in the absence of nitrogen, suggesting a role for WRKY1 in energy conservation. In both light- and nitrogen-dependent processes, the W-box was the most enriched cis-regulatory element in the promoter regions of the WRKY1-responsive genes, suggesting the potential direct transcriptional regulation of these genes by WRKY1. Further analysis identified a reciprocal pattern for the expression of genes regulated by light and nitrogen treatment, i.e., genes repressed by WRKY1 shared a significant overlap with genes repressed by light treatment, but induced by nitrogen treatment, suggesting the role of WRKY1 as an integrator of light and nitrogen signalling pathways.
Having demonstrated that WRKY1 is a regulatory node in the A. thaliana light and nitrogen interaction network, Heerah et al. (2019) performed combined light and nitrogen treatments on wild-type and wrky1 plants to decipher the transcriptional crosstalk between these signalling pathways. The response from this combined treatment was different from, but had overlaps with, the response triggered by the individual treatments, suggesting a crosstalk between the two signalling pathways. Of the WRKY1-regulated genes shared between the light and nitrogen pathways, ~80% were found to be unique to the combined treatment, compared to the individual treatments, further suggesting a direct connection between the two signalling pathways.
A phenotypic analysis of wild-type and wrky1 mutants in nitrogen-poor and -rich environments suggests a role for WRKY1 in energy conservation, wherein carbon resources are mobilized in the dark when nitrogen is abundant, and nitrogen metabolism is activated in the dark when nitrogen is limiting. This hypothesis was supported by the increased carbohydrate (sucrose, glucose and fructose) content in nitrogen-deficient plants in low light conditions, and the increased glutamate and tryptophan pools in nitrogen-deficient plants irrespective of light conditions (Heerah et al., 2019). The response of WRKY1 to low energy signalling and the phenotype of wrky1 mutants highlight a functionally non-redundant role for the A. thaliana WRKY1 transcription factor.
In conclusion, this study demonstrates the crucial role of WRKY1 in regulating responses to light and nitrogen signalling in primary metabolic processes like nitrogen uptake and assimilation, as well as secondary metabolic defense response pathways in A. thaliana. The role of WRKY1 in activating genes involved in the recycling of cellular energy resources when light is limiting but nitrogen is abundant, and the upregulation of amino acid metabolism when both light and nitrogen are limiting, is facilitated by the crosstalk between light and nitrogen signalling pathways through the WRKY1 hub.
References
Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci U S A 105: 4939–4944
Heerah AS, Katari M, Penjor R, Coruzzi G (2019) Running head : WRKY1 regulates light and nitrogen signaling pathways TITLE : WRKY1 mediates transcriptional crosstalk between light and nitrogen signaling pathways in Arabidopsis thaliana 1 . Department of Plant Biology , University of Illinois , 1201 W G. Plant Physiol. https://doi.org/10.1104/pp.19.00685
Krapp A, Berthomé R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou JP, Daniel-Vedele F (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282
Krouk G, Tranchina D, Lejay L, Cruikshank AA, Shasha D, Coruzzi GM, Gutiérrez RA (2009) A systems approach uncovers restrictions for signal interactions regulating genome-wide responses to nutritional cues in Arabidopsis. PLoS Comput Biol. doi: 10.1371/journal.pcbi.1000326
Thum KE, Shasha DE, Lejay L V, Coruzzi GM (2003) Light- and Carbon-Signaling Pathways. Modeling Circuits of Interactions. Plant Physiol 132: 440–452
Ülker B, Somssich IE (2004) WRKY transcription factors: From DNA binding towards biological function. Curr Opin Plant Biol 7: 491–498