What We’re Reading: October 12
Guest editor: Magdalena Julkowska 
Magdalena is a PostDoc at King Abdullah University for Science and Technology (KAUST, Saudi Arabia) working with Prof. Mark Tester. Her main interests are (1) salt-induced changes in root-to-shoot ratio in Arabidopsis, (2) study the expression patterns in plants with enhanced sodium accumulation in their roots and (3) development of tools for data analysis based on R/Shiny. She is passionate about capturing plant architecture using simple models, understanding plant physiology and salinity tolerance.
Review – Getting to the Roots: A Developmental Genetic View of Root Anatomy and Function From Arabidopsis to Lycophytes 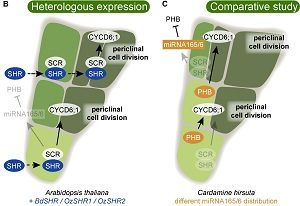
Plant roots are essential organs for water and nutrient uptake, and aboveground biomass support. While these organs are important for many plant functions, the evolutionary history of the root is still unclear. The fossil record suggests that roots evolved in both the lycophyte (clubmosses and their allies) and the euphyllophyte (ferns and seed plants) lineages. Lycophyte and euphyllophyte roots share a number of similarities, such as indeterminate growth due to a root apical meristem, root hair production and the presence of a specialized endodermal layer around the vascular stele. These unique traits must have evolved independently in the lycophyte and euphyllophyte lineages. However, it is unclear how these apparently analogous tissues arose, as there are very few studies that elucidate the genetic and molecular regulation of lycophyte and fern roots. Thus, Augstein and Carlsbecker review key regulatory networks of root development in the angiosperm Arabidopsis and discuss the importance of comparative analyses of root anatomy and morphology of extant and extinct species. This review will aid researchers who are interested in elucidating root evolution through the increased understanding of gene regulation networks and gene function of lycophyte and euphyllophyte roots. (Summary by Julia Miller) Front. Plant Sci. 10.3389/fpls.2018.01410
Review – Cellular basis of growth in plants: geometry matters ($) 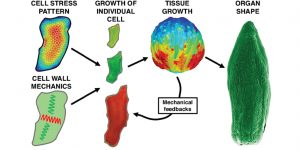
Plants exhibit various forms which are determined by the individual cells. The diversity of cell shapes within a single organism is astounding and results from the interactions between the pressure generated by the cell and surrounding tissue as well as heterogeneities in the cell wall composition. As the pressure within the cell is comparable with the pressure within the champagne bottle, the cell wall is determining the growth direction. When cell wall composition is uniform, the stress distribution depends on cell geometry. Kierzkowski & Routier discuss in their review how cell and organ shapes arise based on cell geometry and tissue context. In the future, integrating life-imaging, mechanical measurements and modeling will provide a better understanding of the regulation of cell shape and organ growth. (Summary by Magdalena Julkowska) Curr. Op in Plant Bio. 10.1016/j.pbi.2018.09.008
The xerobranching response represses lateral root formation when roots are not in contact with water ($)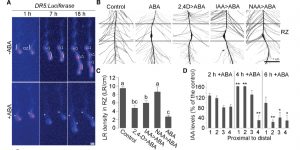
Roots navigate through the soil, foraging for water and nutrients. Orman-Ligeza et al. observed that lateral root development is repressed when the roots are growing through the soil air spaces. Exposure to water deficit induced transcriptome reprogramming in barley roots of genes involved in many hormone pathways. The reduced lateral root formation was also occurring in roots transiently exposed to abscisic acid (ABA), while ABA-insensitive mutants did not show changes in lateral root formation. By using auxin reporters, the authors discovered that reduced lateral root formation is due to decreased auxin levels at the root tip resulting in reduced initiation of the lateral root founder cells. Treatment of the roots with auxin restored lateral root development. How ABA modulates auxin accumulation remains to be determined in future studies. (Summary by Magdalena Julkowska) Curr. Biol. 10.1016/j.cub.2018.07.074
Complete substitution of a secondary cell wall with primary cell wall in Arabidopsis ($)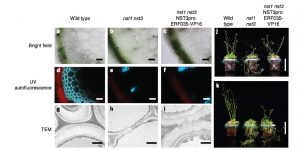
Plant cell walls are important for terrestrial lifestyle, providing support and directing the plant growth. The primary and secondary cell walls differ in their chemical composition and flexibility. Secondary cell walls are less perceptive to industrial degradation processes and therefore pose problems in turning the plant biomass into simpler sugars. To identify new regulatory components of cell wall synthesis Sakamoto et al. screened nts1/nts3 mutants, lacking secondary cell walls in the interfascicular fibre cells, complemented with selected 300 transcription factors under the NTS3 promoter. The nts1/nts3 phenotype was complemented with ERF035 construct, resulting in the thicker cell walls without lignin, which is characteristic to the secondary cell walls. ERF035 was observed to regulate the transcription of primary cell wall biosynthesis genes and was directly interacting with the promoter elements of Cellulose Synthase (CESA1). Although the composition of thickened walls in ERF035 lines resembled primary cell walls, the higher degree of cellulose crystallization and polymerization was observed. (Summary by Magdalena Julkowska) Nature Plants 10.1038/s41477-018-0160-4
Abscisic acid-independent stomatal CO2 signal transduction pathway and convergence of CO2 and ABA signaling downstream of OST1 kinase ($)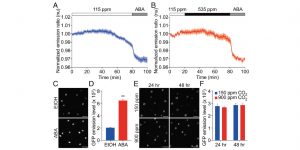
The global increase in CO2 concentrations results in the reduced opening of the stomata. Although reduction in the stomatal opening in response to drought relies on abscisic acid (ABA), less is known about the mechanism controlling stomatal aperture in response to elevated CO2. Hsu et al. examined stomatal CO2 responses in Arabidopsis using mutant lines in ABA biosynthesis and perception. While ABA signal transduction accelerated stomatal responses, mutants in ABA perception were still able to respond to increased CO2 levels. The activation of SnRK2.6/OST1 kinase was observed during the ABA treatment, but not in response to elevated CO2. The guard cell S-type anion channels were activated by CO2 in the ABA-insensitive mutants, suggesting an ABA-independent pathway for CO2-induced stomatal closure. (Summary by Magdalena Julkowska) PNAS 10.1073/pnas.1809204115
FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana ($)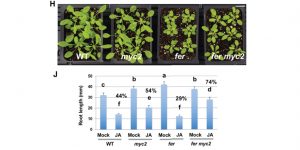
Receptor-like kinase FERONIA plays important role in the regulation of plant growth as well as interactions with the environment. In order to understand how FER affects plant growth Guo et al. examined global transcriptome changes in fer mutant. The authors observed that genes up-regulated by stress hormones, including jasmonic (JA) and abscisic acid (ABA), were also highly expressed in fer mutant. Constitutive activation of JA resulted in higher pathogen susceptibility of fer, which was partially suppressed in myc2 fer mutant. FER was observed to interact and phosphorylate MYC2, leading to MYC2 destabilization and increased plant immunity. The interaction between FER and its peptide ligand RALF23 was observed to have a negative effect on plant growth and immunity by stabilization of MYC2. (Summary by Magdalena Julkowska) Curr. Biol. 10.1016/j.cub.2018.07.078
The genetic architecture of colonization resistance in Brachypodium distachyon to non-adapted stripe rust (Puccinia striiformis) isolates 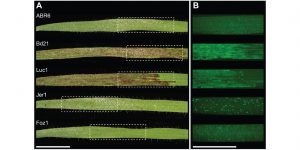
The host range of a pathogen depends on its ability to overcome various plant defense barriers and successfully complete its life cycle. Although host “jumps” are considered rare, the pathogens are able to infect plants other than their usual host with varying degree of success. Yellow stripe rust (P. striiformis) is usually feasting on wheat and barley, and can colonize but is not able to complete its life cycle on the wild grass Brachypodium distachyon. Bettgenhaeuser et al. aimed to understand the genetic architecture restricting colonization by yellow stripe rust by screening three B. distachyon mapping populations. Two major effect QTLs conferring Yellow rust resistance (Yrr1 and Yrr3) conferred resistance to diverse yellow rust isolates. Yrr3 contained immune receptors from the nucleotide-binding leucine-rich repeat (NB-LRR) family, which are also found in the adapted host systems. This study suggests that the resistance to adapted and non-adapted pathogens might not be that different. (Summary by Magdalena Julkowska) PLOS Gen. 10.1371/journal.pgen.1007637
A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis. 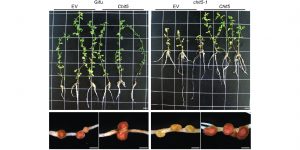
Nitrogen-fixing bacteria produce species-specific chitin-like molecules, Nod factors, which induce nodule development and infection thread formation in the host plant, aiding microbial infection. Malolepszy et al. performed a detailed study of symbiotic defective mutants in lotus (chit5), where the nodule primordia do not develop into nodules after inoculation with M.loti. CHIT5 was observed to function in infection thread development and branching within nodules, which is required for the establishment of fully functional nitrogen-fixing symbiosis. Chit5 was actively transcribed in all cells of uninoculated roots and early stages of infection and decreased within fully functional nodules. The secreted CHIT5 contributed to hydrolysis of a Nod factor R7A, but the hydrolysis was limited to fully compatible Nod-factors – ensuring symbiote-specific signaling. CHIT5 dependent modulation of Nod factor levels within nodules is critical for transition to full symbiosis with nitrogen-fixing bacteria. (Summary by Magdalena Julkowska) eLIFE 10.7554/eLife.38874
The number of meiotic double-strand breaks influences crossover distribution in Arabidopsis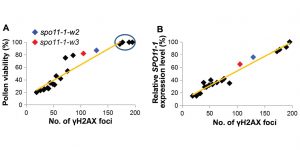
The new genetic variation is generated by recombination of the chromosome during meiotic cell divisions. The programmed double-strand breaks are initiated by DNA Topoisomerase VI-a Subunit (SPO11). As the repair of the double-strand breaks is tightly controlled, the number of breaks usually exceeds the number of crossover events. However, the mechanisms controlling recombination dynamics are largely unknown. Xue et al. studied incomplete complementation mutants of spo11 and observed that decreasing numbers of cross-overs were proportional to the double-strand breaks. Interestingly, change in cross-over distribution observed in SPO11 lines was observed primarily in pericentromeric chromosome regions. The chromatin modification patterns are likely contributing to the crossover distribution. This study provides an example of how genetic diversity can be modulated by the expression of one gene. (Summary by Magdalena Julkowska) The Plant Cell 10.1105/tpc.18.00531
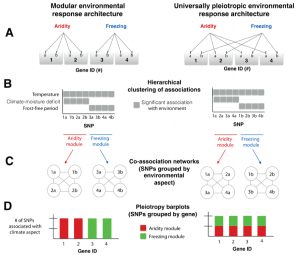
Modularity of genes involved in local adaptation to climate despite physical linkage
Studying local adaptation is a tricky as the environmental parameters that we measure might affect the organism’s fitness. When local adaptation is driven by complex and non-covarying stresses, it is predicted that loci with similar pleiotropic effect are favored. Lotterhos et al. developed a framework for characterizing genetic architecture of adaptation by using joint inference of loci associated with different environmental factors. The framework was used for studying the genetic architecture of lodgepole pine (Pinus contorte) and identified many clusters of SNPs associated with distinct environments, including aridity and freezing. The physical linkage among loci adapting to different aspects of climate was discovered, providing insight into ecological pressures favoring the evolution of genetic modules by natural selection. (Summary by Magdalena Julkowska) Genome Biol. 10.1186/s13059-018-1545-7
Digital imaging combined with genome-wide association mapping links loci to plant-pathogen interaction traits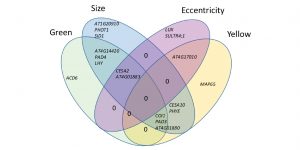
Resistance to plant pathogens is often studied as a qualitative trait than quantitative, focusing on the lesion size and pathogen numbers. However, resistance to generalist plant pathogens, such as Botrytis cinerea, is known to involve multiple genes. Fordyce et al. used high-throughput phenotyping for quantification of Botrytis infection in Arabidopsis. The authors studied color, shape, and size of the lesion and used 96 Arabidopsis accessions to identify genetic components of those alternative disease-related traits. Lesion size and color corresponded to jasmonic acid receptor COI1, cellulose synthase genes as well as genes related to light signaling. Lesion shape was genetically distinct from the other traits and corresponded to genes involved in signal transduction and sulfur transport. The authors propose the use of quantitative methods for studying other non-specialist pathogen infection in combination with forward genetics to identify new breeding target for biotic stress resistance. (Summary by Magdalena Julkowska) Plant Phys. 10.1104/pp.18.00851
A gene‐stacking approach to overcome the trade‐off between drought stress tolerance and growth in Arabidopsis
In the face of increasing incidence of drought events, developing drought-tolerant plants becomes urgent matter. However, the increase in drought tolerance often coincides with the significant reduction of plant size, as in the case of overexpressing DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 1A (DREB1A). Kudo et al. combined DREB1A with genes promoting plant growth, including gibberellin biosynthesis enzyme GA REQUIRING5 (GA5) and PHYTOCHROME INTERACTING FACTOR4 (PIF4) which regulates cell growth. Double overexpression lines developed larger rosettes compared to DREB1A lines, but the drought tolerance was maintained only in GA5 DREB1A lines, due to downregulation of DREB1A expression in PIF4 DREB1A lines. The detailed analysis of the double overexpression lines revealed upregulation of tricarboxylic acid cycle, increased metabolic rates and altered homeostasis of auxin and gibberellin. This study exemplifies that gene stacking of drought resistance and growth genes can be successfully applied in molecular breeding for enhanced drought tolerance. (Summary by Magdalena Julkowska) Plant J 10.1111/tpj.14110
Rhizosphere microbiota structure alters to enable wilt resistance in tomato ($)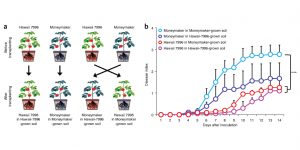
Plants are frequently attacked by pathogens, but the pathogen-resistance is often attributed to the host. While interactions between the plant and its microbiota are recognized for their role in plant growth, their role in pathogen resistance is unknown. Kwak et al. studied the effect of soil-borne pathogen Ralstonia solanacearum causing bacterial wilt disease in tolerant and susceptible tomato varieties, Hawaii 7996 and Moneymaker respectively. The metagenomics analyses revealed that Flavobacteriacea species were dominant in the Hawai 7995 rhizosphere microbiome and identified TRG1-10 strain to reduce wilting due to Ralstonia infection in Moneymaker. This study shows that disease-resistant tomatoes may recruit bacterial allies to protect themselves from soil-borne pathogens and that disease resistance relies on the holobiome of the hosts and its microbial allies. (Summary by Magdalena Julkowska) Nature Biotech. 10.1038/nbt.4232



