What We’re Reading: July 13th
Review. Genetically encoded biosensors in plants: Pathways to discovery ($)
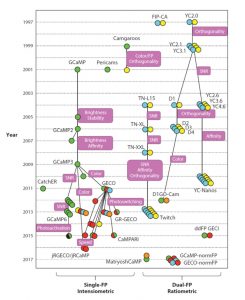 Genetically-encoded biosensors are produced from genes, and provide a specific readout (usually fluorescence or luminescence) of the amount and distribution of a compund of interest (the analyte). We’ve all see data obtained from genetically encoded biosensors, such as the widely-used cameleon family of calcium sensor, but did you ever think about what it takes to develop these essential tools? Walia et al. provide a comprehensive review of the key features that make these tools useful and how they are developed and optimized (including an intriguing “evolutionary history” of biosensors). They also detail the families of analytes that biosensors have been developed towards, including hormones and nutrients, and how they’ve informed our understanding of plant biology. Interestingly, the authors indicate that biosensors developed for use in other organisms usually are functional in plants, lowering the barrier for their application. (Summary by Mary Williams) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-042817-040104
Genetically-encoded biosensors are produced from genes, and provide a specific readout (usually fluorescence or luminescence) of the amount and distribution of a compund of interest (the analyte). We’ve all see data obtained from genetically encoded biosensors, such as the widely-used cameleon family of calcium sensor, but did you ever think about what it takes to develop these essential tools? Walia et al. provide a comprehensive review of the key features that make these tools useful and how they are developed and optimized (including an intriguing “evolutionary history” of biosensors). They also detail the families of analytes that biosensors have been developed towards, including hormones and nutrients, and how they’ve informed our understanding of plant biology. Interestingly, the authors indicate that biosensors developed for use in other organisms usually are functional in plants, lowering the barrier for their application. (Summary by Mary Williams) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-042817-040104
Review. Actions of plant Argonautes: Predictable or unpredictable? ($)
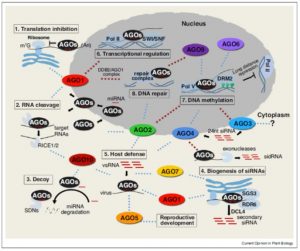 The Arabidopsis genome encodes nine Argonaute proteins and an AGO pseudogene. The nine functional proteins fall into three clades based on sequence. Ma and Zhang update what we know about these proteins. It turns out, the simple assumptions made early on have not entirely been borne out. For example, AGO1 helps miRNAs carry out their functions (slicing and translational repression), but it also has roles in the nucleus, as a transcriptional repressor and transactivator. The finding that this and other AGO proteins can have multiple roles raises questions about how these functions are coordinated. Further, it indicates that the classification of AGO function based on phylogeny needs to be re-evaluated. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2018.05.007
The Arabidopsis genome encodes nine Argonaute proteins and an AGO pseudogene. The nine functional proteins fall into three clades based on sequence. Ma and Zhang update what we know about these proteins. It turns out, the simple assumptions made early on have not entirely been borne out. For example, AGO1 helps miRNAs carry out their functions (slicing and translational repression), but it also has roles in the nucleus, as a transcriptional repressor and transactivator. The finding that this and other AGO proteins can have multiple roles raises questions about how these functions are coordinated. Further, it indicates that the classification of AGO function based on phylogeny needs to be re-evaluated. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2018.05.007
Review. Brachypodium: A monocot grass model system for plant biology
 Brachypodium distachyon is an annual C3 grass that has become an important model species. Scholthof et al. review the genetic tools and resources (sequences and mutants) as well as robust protocols for transformation that have been developed for it. The presence of wild and perennial species within the Brachypodium genus provides additional value. The authors summarize some of the hot topics being addressed through this model organism, including monocot cell wall biochemistry and biofuel production, control of flowering time, root biology, and abiotic and biotic stress responses (usefully, a list of microbes and invertebrates that affect Brachypodium is given). The article also provides links to the relevant resource websites and databases. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00083
Brachypodium distachyon is an annual C3 grass that has become an important model species. Scholthof et al. review the genetic tools and resources (sequences and mutants) as well as robust protocols for transformation that have been developed for it. The presence of wild and perennial species within the Brachypodium genus provides additional value. The authors summarize some of the hot topics being addressed through this model organism, including monocot cell wall biochemistry and biofuel production, control of flowering time, root biology, and abiotic and biotic stress responses (usefully, a list of microbes and invertebrates that affect Brachypodium is given). The article also provides links to the relevant resource websites and databases. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00083
Review. Out of shape during stress: a key role for auxin
 The plant hormone auxin is a major player in determining root growth and architecture, but we are just starting to understand how auxin distribution is altered by abiotic stresses. Kover et al. discuss how the “upside-down fountain” of auxin in the root is affected by abiotic stress conditions. Auxin transporters not only contribute to the auxin gradient along the root, but also control auxin distribution within the cell, for example by sequestering the auxin in the vacuole or ER and so restricting its accessibility for nuclear signaling. Also, the decrease of the cytoplasmic and apoplastic pH restricts auxin mobility under saline conditions. Although the majority of auxin is believed to be produced in the shoot, genes involved in auxin synthesis are locally upregulated in the root under stress, contributing to the auxin pool and sustaining the growth of the main and lateral roots. The authors argue for the integration of auxin metabolism and local regulation of auxin biosynthesis into mathematical models to better understand how auxin modulates root development under stress conditions. (Summary by Magdalena Julkowska) Trends Plant Sci. 10.1016/j.tplants.2018.05.011
The plant hormone auxin is a major player in determining root growth and architecture, but we are just starting to understand how auxin distribution is altered by abiotic stresses. Kover et al. discuss how the “upside-down fountain” of auxin in the root is affected by abiotic stress conditions. Auxin transporters not only contribute to the auxin gradient along the root, but also control auxin distribution within the cell, for example by sequestering the auxin in the vacuole or ER and so restricting its accessibility for nuclear signaling. Also, the decrease of the cytoplasmic and apoplastic pH restricts auxin mobility under saline conditions. Although the majority of auxin is believed to be produced in the shoot, genes involved in auxin synthesis are locally upregulated in the root under stress, contributing to the auxin pool and sustaining the growth of the main and lateral roots. The authors argue for the integration of auxin metabolism and local regulation of auxin biosynthesis into mathematical models to better understand how auxin modulates root development under stress conditions. (Summary by Magdalena Julkowska) Trends Plant Sci. 10.1016/j.tplants.2018.05.011
How to identify the substrates of ATP-binding cassette (ABC) transporters
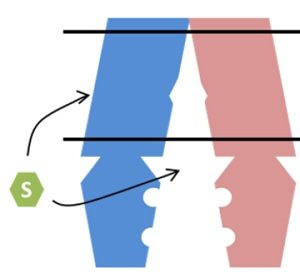 Plant genomes encode many ATP-binding cassette (ABC) transporters, which mediate the transport of many different substrates involved in development, nutrition and stress responses. Mutants and transgenic plants have been used to understand the in planta roles, yet the identity of substrates transported by many ABC proteins remain unknown. Since these proteins are able to transport many structurally unrelated substrates, and since the identification of substrates is often performed indirectly, substrate identification has been difficult. This review by Lefèvre and Boutry discusses ABC transporters that have unambiguously identified substrates, as well as heterologous and homologous expression systems used to study ABC transporters. They also discuss how current research is elucidating ABC transporter structures, bidirectionality of transport, and the role of post-translational modifications. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.18.00325
Plant genomes encode many ATP-binding cassette (ABC) transporters, which mediate the transport of many different substrates involved in development, nutrition and stress responses. Mutants and transgenic plants have been used to understand the in planta roles, yet the identity of substrates transported by many ABC proteins remain unknown. Since these proteins are able to transport many structurally unrelated substrates, and since the identification of substrates is often performed indirectly, substrate identification has been difficult. This review by Lefèvre and Boutry discusses ABC transporters that have unambiguously identified substrates, as well as heterologous and homologous expression systems used to study ABC transporters. They also discuss how current research is elucidating ABC transporter structures, bidirectionality of transport, and the role of post-translational modifications. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.18.00325
Lateral root priming synergistically arises from root growth and auxin transport dynamics
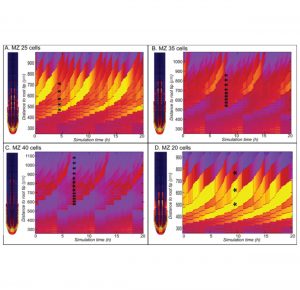 Root system architecture is primarily determined by branching of the lateral roots. The position of a new lateral root is determined by oscillating auxin concentrations in the main root meristem. To understand the nature of auxin oscillations Van den Berg and ten Tusscher developed a multiscale root model integrating realistic root tip architecture, auxin reflux from the lateral root cap, and cell growth dynamics. In elongating vascular cells, the surface occupied by polar auxin efflux proteins (PINs) decreases relative to non-polar auxin uptake, creating a high auxin uptake zone that is fed by auxin reflux from lateral root cap cells. Together with small differences in the size of cells produced by the meristem, this efffect is sufficient to create regular auxin oscillations. According to the model, meristem size has a negative effect on the density of oscillations and thus the frequency of primed sites. The new model suggests that the reduction in main root growth and increase in lateral root density, as observed under phosphate starvation, can be explained by one and the same change in root dynamics. (Summary by Magdalena Julkowska) bioRxiv 10.1101/361709
Root system architecture is primarily determined by branching of the lateral roots. The position of a new lateral root is determined by oscillating auxin concentrations in the main root meristem. To understand the nature of auxin oscillations Van den Berg and ten Tusscher developed a multiscale root model integrating realistic root tip architecture, auxin reflux from the lateral root cap, and cell growth dynamics. In elongating vascular cells, the surface occupied by polar auxin efflux proteins (PINs) decreases relative to non-polar auxin uptake, creating a high auxin uptake zone that is fed by auxin reflux from lateral root cap cells. Together with small differences in the size of cells produced by the meristem, this efffect is sufficient to create regular auxin oscillations. According to the model, meristem size has a negative effect on the density of oscillations and thus the frequency of primed sites. The new model suggests that the reduction in main root growth and increase in lateral root density, as observed under phosphate starvation, can be explained by one and the same change in root dynamics. (Summary by Magdalena Julkowska) bioRxiv 10.1101/361709
Expression of FLOWERING LOCUS C differentiates summer and winter biotypes of Camelina sativa
 Camelina sativa is a member of the Brassicaceae family that is grown as an oilseed crop. It has summer and winter flowering biotypes, the latter of which requires vernalization to induce flowering. These two biotypes allow for double-cropping, and the winter-flowering biotype can also be used as a cover crop. To investigate the genetic basis for the different flowering requirements in this species, Anderson et al. carried out whole-genome and RNA sequencing studies of the two biotypes. The Camelina sativa genome encodes three loci encoding the floral repressor FLC, one of which is expressed at much higher levels in the veranlization-requiring plants. Interestingly, the Chromosome 20 locus appears to contain three copies of the FLC gene, one of which has a single-nucleotide deletion that would result in a frame shift and non-functional protein. Transcripts corresponding to this frame-shift variant are more highly expressed in the summer flowering species. (Summary by Mary Williams) (Image by Fornax) Plant Direct J. 10.1002/pld3.60
Camelina sativa is a member of the Brassicaceae family that is grown as an oilseed crop. It has summer and winter flowering biotypes, the latter of which requires vernalization to induce flowering. These two biotypes allow for double-cropping, and the winter-flowering biotype can also be used as a cover crop. To investigate the genetic basis for the different flowering requirements in this species, Anderson et al. carried out whole-genome and RNA sequencing studies of the two biotypes. The Camelina sativa genome encodes three loci encoding the floral repressor FLC, one of which is expressed at much higher levels in the veranlization-requiring plants. Interestingly, the Chromosome 20 locus appears to contain three copies of the FLC gene, one of which has a single-nucleotide deletion that would result in a frame shift and non-functional protein. Transcripts corresponding to this frame-shift variant are more highly expressed in the summer flowering species. (Summary by Mary Williams) (Image by Fornax) Plant Direct J. 10.1002/pld3.60
Aquatic fern genomes provide insight into land plant evolution and symbiosis
 Land plants evolved from freshwater charophytic algae over ~450 million years ago and have since diverged into the plethora of embryophyte genera that we see today. Genomics efforts have classically focused on key angiosperm species representing experimental model systems and/or agriculturally important crops, yet an increasing amount of attention is now being given to earlier diverging plant lineages. In a keystone study published in Nature Plants, Li et al. describe the successful assembly and analysis of two aquatic fern genomes (Salvina cucullata and Azolla filiculoides). The authors discuss recent whole genome duplication events in ferns and the likely acquisition of insecticide-related genes by horizontal gene transfer from bacteria. In addition, in-depth phylogenetic analyses demonstrated that Azolla maintains a unique symbiotic relationship with cyanobacteria despite lacking gene families essential for root-nodule (bacteria) and arbuscular mycorrhizal (fungal) symbiosis. Collectively, this work sets the stage for future comparative genomics analyses that will address questions pertaining to the evolution of plant-cyanobacteria symbiosis and other key plant traits. (Summary by Phil Carella) Nature Plant 10.1038/s41477-018-0188-8
Land plants evolved from freshwater charophytic algae over ~450 million years ago and have since diverged into the plethora of embryophyte genera that we see today. Genomics efforts have classically focused on key angiosperm species representing experimental model systems and/or agriculturally important crops, yet an increasing amount of attention is now being given to earlier diverging plant lineages. In a keystone study published in Nature Plants, Li et al. describe the successful assembly and analysis of two aquatic fern genomes (Salvina cucullata and Azolla filiculoides). The authors discuss recent whole genome duplication events in ferns and the likely acquisition of insecticide-related genes by horizontal gene transfer from bacteria. In addition, in-depth phylogenetic analyses demonstrated that Azolla maintains a unique symbiotic relationship with cyanobacteria despite lacking gene families essential for root-nodule (bacteria) and arbuscular mycorrhizal (fungal) symbiosis. Collectively, this work sets the stage for future comparative genomics analyses that will address questions pertaining to the evolution of plant-cyanobacteria symbiosis and other key plant traits. (Summary by Phil Carella) Nature Plant 10.1038/s41477-018-0188-8
Scientists on Twitter: Preaching to the choir or singing from the rooftops?
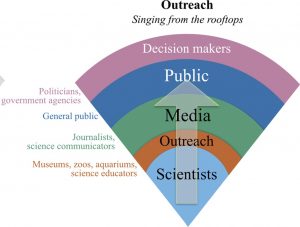 Science communication is as old as science itself, reaching even best-selling levels with some extraordinary examples such as “On the Origin of Species” by Charles Darwin or “A Brief History of Time: From the Big Bang to Black Holes” by Stephen Hawking. Currently, and thanks to social media, communication including science communication is incredibly accelerated. However, the real reach of scientists in social media in general, and Twitter in particular, is still unknown both from a quantitative and qualitative point of view. In Cotê & Darling (2018), the researchers shed light on this matter by studying the real reach of 110 Twitter accounts managed by individual scientists. They found that scientists are predominantly followed by other scientist (55%), however the more followers the account had (beyond 1000) the more diverse those followers became, confirming that tweeting has potential to disseminate scientific content to the general public. According to the authors “These results should encourage scientists to invest in building a social media presence for scientific outreach”. (Summary by Isabel Mendoza) Facets Journal 10.1139/facets-2018-0002
Science communication is as old as science itself, reaching even best-selling levels with some extraordinary examples such as “On the Origin of Species” by Charles Darwin or “A Brief History of Time: From the Big Bang to Black Holes” by Stephen Hawking. Currently, and thanks to social media, communication including science communication is incredibly accelerated. However, the real reach of scientists in social media in general, and Twitter in particular, is still unknown both from a quantitative and qualitative point of view. In Cotê & Darling (2018), the researchers shed light on this matter by studying the real reach of 110 Twitter accounts managed by individual scientists. They found that scientists are predominantly followed by other scientist (55%), however the more followers the account had (beyond 1000) the more diverse those followers became, confirming that tweeting has potential to disseminate scientific content to the general public. According to the authors “These results should encourage scientists to invest in building a social media presence for scientific outreach”. (Summary by Isabel Mendoza) Facets Journal 10.1139/facets-2018-0002



