What We’re Reading: January 25th
Special Issue: Genome to Phenome (Plant Journal)
 It’s January, which seems to be a month of weather extremes for many “temperate” parts of the globe. Here’s an excellent collection to curl up with as you try to stay warm or cool and out of the elements. The issue includes free-to-access reviews focused on using genomic data (e.g., genome-wide associate studies, GWAS), to identify genes or loci that correlate with various phenotypes. On the genome end, the articles discuss tools and approaches, especially focused on crop plants or plants with large and complex genomes. On the phenome side, articles cover metabolic traits, salt tolerance, photosynthetic efficiency, cell walls and more. This would be a great reading list for a journal club, especially for students! (Summary by Mary Williams) https://onlinelibrary.wiley.com/toc/1365313x/2019/97/1
It’s January, which seems to be a month of weather extremes for many “temperate” parts of the globe. Here’s an excellent collection to curl up with as you try to stay warm or cool and out of the elements. The issue includes free-to-access reviews focused on using genomic data (e.g., genome-wide associate studies, GWAS), to identify genes or loci that correlate with various phenotypes. On the genome end, the articles discuss tools and approaches, especially focused on crop plants or plants with large and complex genomes. On the phenome side, articles cover metabolic traits, salt tolerance, photosynthetic efficiency, cell walls and more. This would be a great reading list for a journal club, especially for students! (Summary by Mary Williams) https://onlinelibrary.wiley.com/toc/1365313x/2019/97/1
Review: Green Bioplastics as part of a circular bioeconomy ($)
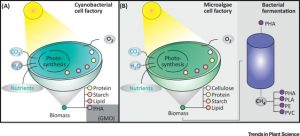 I feel guilty about using plastics but it’s hard to break free; they are so integral to a modern lifestyle. However, the problem of plastic pollution is frightening. About 80% of manufactured plastic ends up in landfills or the natural environment. It’s appalling to learn that the “Great Pacific Garbage Patch” covers 1.6 million km2. That’s why I am encouraged by articles like this from Karan and Funk et al., on the potential for green, biodegradable plastics; I want guilt-free plastics. This review focuses on the potential for green plastic production by cyanobacteria and green algae (to avoid competition for arable land with food-producing plants). The article describes different types of polymers that can be derived from photosynthetic cells, their applications, and potential market values. It also discusses the behind-the-scences chemistry that needs to take place both in terms of synthesis and biorefining to produce clean, green plastics. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.11.010
I feel guilty about using plastics but it’s hard to break free; they are so integral to a modern lifestyle. However, the problem of plastic pollution is frightening. About 80% of manufactured plastic ends up in landfills or the natural environment. It’s appalling to learn that the “Great Pacific Garbage Patch” covers 1.6 million km2. That’s why I am encouraged by articles like this from Karan and Funk et al., on the potential for green, biodegradable plastics; I want guilt-free plastics. This review focuses on the potential for green plastic production by cyanobacteria and green algae (to avoid competition for arable land with food-producing plants). The article describes different types of polymers that can be derived from photosynthetic cells, their applications, and potential market values. It also discusses the behind-the-scences chemistry that needs to take place both in terms of synthesis and biorefining to produce clean, green plastics. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.11.010
Reviews: The regulation of cellulose biosynthesis in plants, and secondary cell wall biosynthesis
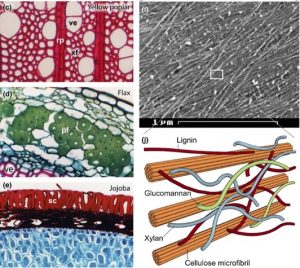 Plant cell walls are complex and somewhat difficult to study, but also idespensible for plants and central to many of the ways that people use plants (from wood to fibers to fuels). Two new articles review diverse aspects of plant cell wall synthesis. Polko and Kieber (Plant Cell 10.1105/tpc.18.00760 $) focus on the regulation of cellulose production and the structure, function and regulation of the cellulose synthase complex. The also review new insights into the intracellular trafficking of cellulose-synthase complexes and its contribution to cell wall sythesis, and the mechanisms behind cell-wall integrity sensing and maintenance. The review by Zhong et al. (New Phytol. 10.1111/nph.15537) focuses on secondary cell wall synthesis. Secondary walls are rigid, form after cell elongation ceases, and strengthen specialized cells including xylem tracheids and vessels. The review highlights insights from genetics and biochemistry into the synthesis of wall components including cellulose, xylan, glucomannan and lignin. Knowledge of cell wall synthesis can be leveraged towards the development of cell walls optimized for specific purposes. (Summary by Mary Williams)
Plant cell walls are complex and somewhat difficult to study, but also idespensible for plants and central to many of the ways that people use plants (from wood to fibers to fuels). Two new articles review diverse aspects of plant cell wall synthesis. Polko and Kieber (Plant Cell 10.1105/tpc.18.00760 $) focus on the regulation of cellulose production and the structure, function and regulation of the cellulose synthase complex. The also review new insights into the intracellular trafficking of cellulose-synthase complexes and its contribution to cell wall sythesis, and the mechanisms behind cell-wall integrity sensing and maintenance. The review by Zhong et al. (New Phytol. 10.1111/nph.15537) focuses on secondary cell wall synthesis. Secondary walls are rigid, form after cell elongation ceases, and strengthen specialized cells including xylem tracheids and vessels. The review highlights insights from genetics and biochemistry into the synthesis of wall components including cellulose, xylan, glucomannan and lignin. Knowledge of cell wall synthesis can be leveraged towards the development of cell walls optimized for specific purposes. (Summary by Mary Williams)
Review: How to analyse plant phenotypic plasticity in response to a changing climate
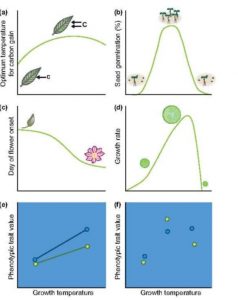 Phenotypic plasticity, or the ability to change in response to the environment, is one of the most characteristic (and to me, endearing) qualities of plants. As Arnold et al. observe, it is also one of the most important in terms of future climate change; phenotypic plasticity will mean the difference between species that persist and those that perish. But, they also observe, phenotypic plasticity is difficult to quantify. Here, they provide an approach by which to analyze phenotypic plasticity, using reaction norms. In the Supporting Information, they provide a tutorial with instructions and code demonstrating how a random regression mixed model framework can be used to characterize plasticity. (Summary by Mary Williams) New Phytol. 10.1111/nph.15656
Phenotypic plasticity, or the ability to change in response to the environment, is one of the most characteristic (and to me, endearing) qualities of plants. As Arnold et al. observe, it is also one of the most important in terms of future climate change; phenotypic plasticity will mean the difference between species that persist and those that perish. But, they also observe, phenotypic plasticity is difficult to quantify. Here, they provide an approach by which to analyze phenotypic plasticity, using reaction norms. In the Supporting Information, they provide a tutorial with instructions and code demonstrating how a random regression mixed model framework can be used to characterize plasticity. (Summary by Mary Williams) New Phytol. 10.1111/nph.15656
Opinion: How can we boost the impact of publications? Try better writing
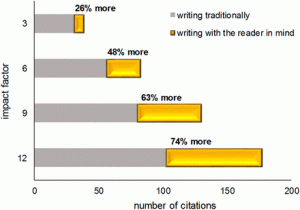 Writing is one of the most important of a scientist’s skills but often overlooked during their training. As Freeling et al. observe, good writing takes time, both to learn and to teach, so what’s the incentive? In this Opinion article, they make an effort to quantify the impact (measured by citation numbers) of “good writing”. They conclude that there is a correlation between better writing and more citations. “Good writing” is of course very hard to quantify, which is the limitation of this study. However, the factors that they have specified as signaling “good writing” are useful to note (after all, they correlate with higher citations!). These include: signposting, consistent language, parallel phrasing, punctuation, and avoidance of noun chunks, as explained in the article. Have a look – we can’t promise that writing better will increase your citation rate, but it can’t hurt! (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1819937116
Writing is one of the most important of a scientist’s skills but often overlooked during their training. As Freeling et al. observe, good writing takes time, both to learn and to teach, so what’s the incentive? In this Opinion article, they make an effort to quantify the impact (measured by citation numbers) of “good writing”. They conclude that there is a correlation between better writing and more citations. “Good writing” is of course very hard to quantify, which is the limitation of this study. However, the factors that they have specified as signaling “good writing” are useful to note (after all, they correlate with higher citations!). These include: signposting, consistent language, parallel phrasing, punctuation, and avoidance of noun chunks, as explained in the article. Have a look – we can’t promise that writing better will increase your citation rate, but it can’t hurt! (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1819937116
Origin and function of the root vascular cambium ($)
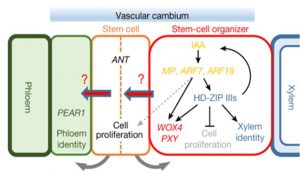 The vascular cambium is a meristematic tissue that is responsible for lateral growth and the continued production of new xylem and phloem; in woody plants, the shoot vascular cambium makes wood. A new pair of papers investigates how the Arabidopsis root vascular cambium forms and how it functions. Through mutant analysis and imaging, the authors unravel several interesting mechanisms for these processes; these include mobile cell-to-cell signals that inform cells about their neighbors and their positions, key roles for hormones, and antagonistically-functioning sets of transcription factors. Thus, a key finding is that although some of the specific players identified are new, the developmental programs involved in vascular cambium development and function follow similar ground rules as those that act in the primary root and shoot meristems. (Miyashima et al. 10.1038/s41586-018-0839-y and Smetana et al. 10.1038/s41586-018-0837-0.) Nature (Summary by Mary Williams)
The vascular cambium is a meristematic tissue that is responsible for lateral growth and the continued production of new xylem and phloem; in woody plants, the shoot vascular cambium makes wood. A new pair of papers investigates how the Arabidopsis root vascular cambium forms and how it functions. Through mutant analysis and imaging, the authors unravel several interesting mechanisms for these processes; these include mobile cell-to-cell signals that inform cells about their neighbors and their positions, key roles for hormones, and antagonistically-functioning sets of transcription factors. Thus, a key finding is that although some of the specific players identified are new, the developmental programs involved in vascular cambium development and function follow similar ground rules as those that act in the primary root and shoot meristems. (Miyashima et al. 10.1038/s41586-018-0839-y and Smetana et al. 10.1038/s41586-018-0837-0.) Nature (Summary by Mary Williams)
Quantitative analysis of auxin sensing in leaf primordia argues against proposed role in regulating leaf dorsoventrality
 This is an interesting paper that touches on several topics. Bhatia et al. address the question of how polarity in leaves is first established: essentially, which comes first, polarity in auxin concentration or polarity in gene expression? The authors conclude that in the earliest leaf primordia, there is no gradient of auxin sensing (suggesting perhaps no gradient in auxin). This finding contradicts several other recent papers, which is one of the reasons it is so interesting. Other studies used different reporters of auxin response, and slightly different methods for analysis, and also looked at slightly older leaves. The dialog between the reviewing editor and the authors, which like all eLIFE papers is included, provides insights into not only leaf polarity but also how scientific knowledge advances incrementally and by refining and sometimes contradicting the findings of others. (Summary by Mary Williams) eLIFE 10.7554/eLife.39298
This is an interesting paper that touches on several topics. Bhatia et al. address the question of how polarity in leaves is first established: essentially, which comes first, polarity in auxin concentration or polarity in gene expression? The authors conclude that in the earliest leaf primordia, there is no gradient of auxin sensing (suggesting perhaps no gradient in auxin). This finding contradicts several other recent papers, which is one of the reasons it is so interesting. Other studies used different reporters of auxin response, and slightly different methods for analysis, and also looked at slightly older leaves. The dialog between the reviewing editor and the authors, which like all eLIFE papers is included, provides insights into not only leaf polarity but also how scientific knowledge advances incrementally and by refining and sometimes contradicting the findings of others. (Summary by Mary Williams) eLIFE 10.7554/eLife.39298
Sunflower pan-genome, evidence for hybridization-altered disease resistance ($)
 Sunflower (Helianthus annuus L.) is an important oil-producing crop, which was domisticated in North America about 4000 years ago with elite varities being developed through the 19th and 20th centuries, narrowing its genetic variation. It retains the ability to hybridize with wild relatives, providing opportunities to introgress desirable traits. Hübner et al. sequenced and analyzed nearly 12 trillion bases (!) from 493 sunflower accessions (landraces, modern cultivars and wild relatives). Through these efforts, they identified the “pan genome” or set of all genes of cultivated sunflower. They also identified genomic regions that had been introgressed from wild relatives, and regions that show an association with resistance to downy mildew disease. These data provide insights into the genetic diversity of cultivated sunflower. (Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0329-0
Sunflower (Helianthus annuus L.) is an important oil-producing crop, which was domisticated in North America about 4000 years ago with elite varities being developed through the 19th and 20th centuries, narrowing its genetic variation. It retains the ability to hybridize with wild relatives, providing opportunities to introgress desirable traits. Hübner et al. sequenced and analyzed nearly 12 trillion bases (!) from 493 sunflower accessions (landraces, modern cultivars and wild relatives). Through these efforts, they identified the “pan genome” or set of all genes of cultivated sunflower. They also identified genomic regions that had been introgressed from wild relatives, and regions that show an association with resistance to downy mildew disease. These data provide insights into the genetic diversity of cultivated sunflower. (Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0329-0
Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome
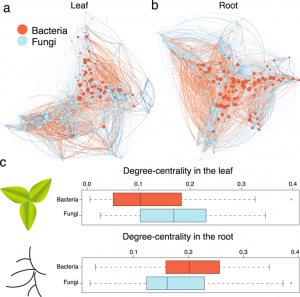 Microbes that live in, on and near plants profoundly affect their growth and survival. Here, Bergelson et al. sequenced the bacterial and fungal microbiomes of Arabidopsis roots (previously they sequenced samples from leaves). The accessions are from all over the world but the sequenced samples are from a single field. Key take-aways are: don’t forget the fungi, they matter too and play an important role in determining the microbiome diversity; the root microbiome is shaped by the genetics of the host; and although many of the genes that underpin the composition of the fungal versus bacterial microbiome are different, a few affect both, including a gene encoding a protein-channel in root hairs and genes involved in cell wall integrity. These data and approaches will help inform efforts to improve plant health through modifying their microbiomes. (Summary by Mary Williams) Sci Reports 10.1038/s41598-018-37208-z
Microbes that live in, on and near plants profoundly affect their growth and survival. Here, Bergelson et al. sequenced the bacterial and fungal microbiomes of Arabidopsis roots (previously they sequenced samples from leaves). The accessions are from all over the world but the sequenced samples are from a single field. Key take-aways are: don’t forget the fungi, they matter too and play an important role in determining the microbiome diversity; the root microbiome is shaped by the genetics of the host; and although many of the genes that underpin the composition of the fungal versus bacterial microbiome are different, a few affect both, including a gene encoding a protein-channel in root hairs and genes involved in cell wall integrity. These data and approaches will help inform efforts to improve plant health through modifying their microbiomes. (Summary by Mary Williams) Sci Reports 10.1038/s41598-018-37208-z
A smut fungus of Brassicaceae plants uses conserved and unique strategies to manipulate perennial hosts
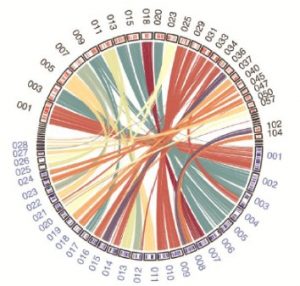 Biotrophic phytopathogens manipulate living hosts for the procurement of nutrients essential for growth and reproduction. It is therefore critical for invading biotrophs to evade or suppress immune responses without impacting overall plant function. In a recent study published in New Phytologist (2019), Courville et al. resolve the genomic and transcriptomic signatures of the smut fungus Thecaphora thlaspeos, which forms long-lasting biotrophic infections with annual and perennial Brassicaceaen hosts. The authors identify a diverse catalog of novel and conserved fungal effectors that mediate biotrophic interactions with plants. The activity of three T. thlaspeos effectors were explored in planta, including a functionally conserved Pep1 protein that partially complemented the virulence defect of a Δpep1 mutant in the corn smut fungus Ustilago maydis. Further, the authors identified a role for the novel TtTue1 effector in suppressing plant immunity, as demonstrated by the enhanced susceptibility of TtTue1-overexpressing Arabidopsis plants to the bacterial pathogen Pseudomonas syringae. Collectively, this work begins to clarify the diversity of effector proteins and virulence strategies used by smut fungi to manipulate the tissues and cells of their living host plants. (Summary by Phil Carella) New Phytol. 10.1111/nph.15692
Biotrophic phytopathogens manipulate living hosts for the procurement of nutrients essential for growth and reproduction. It is therefore critical for invading biotrophs to evade or suppress immune responses without impacting overall plant function. In a recent study published in New Phytologist (2019), Courville et al. resolve the genomic and transcriptomic signatures of the smut fungus Thecaphora thlaspeos, which forms long-lasting biotrophic infections with annual and perennial Brassicaceaen hosts. The authors identify a diverse catalog of novel and conserved fungal effectors that mediate biotrophic interactions with plants. The activity of three T. thlaspeos effectors were explored in planta, including a functionally conserved Pep1 protein that partially complemented the virulence defect of a Δpep1 mutant in the corn smut fungus Ustilago maydis. Further, the authors identified a role for the novel TtTue1 effector in suppressing plant immunity, as demonstrated by the enhanced susceptibility of TtTue1-overexpressing Arabidopsis plants to the bacterial pathogen Pseudomonas syringae. Collectively, this work begins to clarify the diversity of effector proteins and virulence strategies used by smut fungi to manipulate the tissues and cells of their living host plants. (Summary by Phil Carella) New Phytol. 10.1111/nph.15692



