HOS15 Co-regulates Photoperiodic Flowering with the Evening Complex via Transcriptional Repression of GIGANTEA
When animals are stressed by unfavorable environmental conditions, they will typically move to a more favorable location to reduce that stress. Unfortunately for plants, they are quite literally rooted to the ground and thus must manage the stress where they stand. In order to deal with adverse environmental conditions, plants evolved numerous molecular and genetic pathways that become triggered by changes in environmental conditions to withstand that stress (Zhu, 2016).
A plant’s flowering time can be affected by seasonal cues including day length, temperature, and environmental signals, as well as several internal cues. Four distinct pathways, known as the autonomous, gibberellin, photoperiod, and vernalization pathways, regulate flowering time through several common flowering genes (Simpson and Dean, 20002). These pathways have been identified to involve epigenetic factors including DNA methylation, histone modifications, and microRNAs, especially when plants are exposed to abiotic stresses (Yaish et al., 2011). It was previously found that HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 15 (HOS15) mediates histone deacetylation in Arabidopsis (Arabidopsis thaliana) (Zhu et al., 2008). Now, Park et al. show that HOS15 forms a complex with Evening Complex (EC) components, a group of proteins that assist in the regulation of the circadian clock, and the histone deacetylase HDA9 (Park et al., 2018).
 To identify the pathway in which HOS15 functions, the authors utilized high-throughput RNA-seq analysis to identify if any marker genes of specific pathways were differentially expressed in the hos15-2 mutant when compared to wildtype. This analysis showed that the expression of photoperiodic genes such as GIGANTEA (GI), F-BOX1 (FKF1), and CONSTANS (CO) was higher in the hos15-2 mutant compared to wildtype. Marker genes in the vernalization and autonomous pathways, however, remained at wildtype levels. Additionally, expression levels of EARLY FLOWERING3 (ELF3, acts upstream of GI), ELF4 (interacts with ELF3), and several genes encoding PSEUDO RESPONSE REGULATORS (PRRs), including PRR7 and PRR9 (which stabilize CO and promote flowering) were also higher in hos15-2. Taken together, these results suggest that HOS15 is involved in GI-mediated flowering. Analysis of double mutants using HOS15 and the other photoperiodic pathway genes showed the same late flowering phenotype that each of the photoperiod single mutants displayed in long days, suggesting that HOS15 functions upstream of these genes (Park et al., 2018).
To identify the pathway in which HOS15 functions, the authors utilized high-throughput RNA-seq analysis to identify if any marker genes of specific pathways were differentially expressed in the hos15-2 mutant when compared to wildtype. This analysis showed that the expression of photoperiodic genes such as GIGANTEA (GI), F-BOX1 (FKF1), and CONSTANS (CO) was higher in the hos15-2 mutant compared to wildtype. Marker genes in the vernalization and autonomous pathways, however, remained at wildtype levels. Additionally, expression levels of EARLY FLOWERING3 (ELF3, acts upstream of GI), ELF4 (interacts with ELF3), and several genes encoding PSEUDO RESPONSE REGULATORS (PRRs), including PRR7 and PRR9 (which stabilize CO and promote flowering) were also higher in hos15-2. Taken together, these results suggest that HOS15 is involved in GI-mediated flowering. Analysis of double mutants using HOS15 and the other photoperiodic pathway genes showed the same late flowering phenotype that each of the photoperiod single mutants displayed in long days, suggesting that HOS15 functions upstream of these genes (Park et al., 2018).
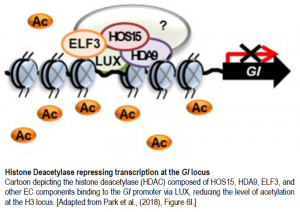
Due to previous findings that HOS15 interacts with histone H4, one of five histones critical in the structure of chromatin, via histone deacetylase complexes (HDACs) to repress cold-induced downstream genes, and the current finding that GI expression is increased in hos15-2 mutants, Park et al. attempted to test if HOS15 typically suppresses GI expression by recruiting HDACs to the GI promoter. The EC regulates GI transcription by binding to a LUX binding site (LBS) in the GI promotor. Using this knowledge, the authors created two amplicons containing two putative LBS, R1 and R3, and a third amplicon without any LBS. Via chromatin immunoprecipitation (ChIP) analysis, interactions between HOS15 and both R1 and R3 regions of the GI promoter were found in wildtype plants, but not in hos15-2 plants. This indicates that HOS15 is recruited to the GI promoter (Park et al., 2018).
The authors’ findings suggest that HOS15 is a transcriptional co-repressor in conjunction with an HDAC, and so thus the authors sought to identify which HDAC HOS15 interacts with. A double mutant analysis of hos15-2 hda9-1 showed similar early flowering seen in the hos15-2 single mutant. ChIP analysis was then used to show that HDA9 interacts with both the R3 region and AcH3 on the GI promotor. The binding of HOS15 to the GI promotor was also reduced in both the hda9-1 and hos15-2 hda9-1 mutants suggesting that both HOS15 and HDA9 regulate flowering via the same pathway. Altogether, the results suggest that HOS15 forms a complex with HDA9, an HDAC, and the EC. This complex then represses GI expression by directly binding to the GI promoter at an LBS (Park et al., 2018).
Park HJ, Baek D, Cha JY, Liao X, Kang SH, McClung CR, Lee SY, Yun DJ and Kim WY (2018). HOS15 interacts with the histone deacetylase HDA9 and the Evening Complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell https://doi.org/10.1105/tpc.18.00721.
Simpson, GG and Dean C (2011). Arabidopsis, the Rosetta stone of flowering time? Science 296: 285-289.
Yaish, MW, Colasanti, J, and Rothstein, SJ (2011). The role of epigenetic processes in controlling flowering time in plants exposed to stress. J. Exp. Bot. 62: 3727–3725
Zhu, JK (2016). Abiotic Stress Signaling and Responses in Plants. Cell 167: 313–324
Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, Hasegawa PM, Bohnert HJ, Shi H, Yun DJ and Bressan RA (2008). Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. USA. 105: 4945–4950.