What We’re Reading: February 9th
Review. Use it or average it: Stochasticity in plant development (Curr. Opin. Plant Biol.)
 In this interesting review, Roeder describes the importance of stochasticity in plant development. She starts off with an explanation: “A process that can be analyzed statistically but not predicted precisely is stochastic. Stochasticity does not imply the absence of regulation, just that the regulation does not cause an exactly determined output in all cases.” She then goes on to illustrate (using a variety of model systems) how patterns can be generated through stochastic fluctuations, as well as how stochasticity contributes to robustness. It’s common when studying plant development to want to ignore its inherent variability, but as this review reminds us these unpredictable events can be important determinants of the developmental outcomes. Curr. Opin. Plant Biol. (Summary by Mary Williams) 10.1016/j.pbi.2017.07.010
In this interesting review, Roeder describes the importance of stochasticity in plant development. She starts off with an explanation: “A process that can be analyzed statistically but not predicted precisely is stochastic. Stochasticity does not imply the absence of regulation, just that the regulation does not cause an exactly determined output in all cases.” She then goes on to illustrate (using a variety of model systems) how patterns can be generated through stochastic fluctuations, as well as how stochasticity contributes to robustness. It’s common when studying plant development to want to ignore its inherent variability, but as this review reminds us these unpredictable events can be important determinants of the developmental outcomes. Curr. Opin. Plant Biol. (Summary by Mary Williams) 10.1016/j.pbi.2017.07.010
Opinion: Plant pathogen effector proteins as manipulators of host microbiomes? (Mol Plant)
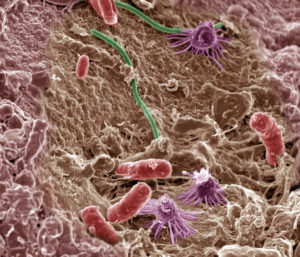 To understand disease development, effector research has mainly focused on the direct interaction of pathogen-derived molecules with plant host targets, or their sensing by surface or intracellular receptors. Recently, attention has turned to the plant microbiome and its key role in maintaining plant health. How microbial communities are manipulated by plant pathogens in sensing and infection of hosts is still largely unexplored. In this Opinion article, Snelders et al. hypothesize the involvement of highly specialized effectors that may act as tools for interaction with beneficial microbes. These microbiome-manipulating effectors may have different transcriptional profiles from other effectors, due to being involved in processes before the actual host contact. Therefore, identifying this interesting set of pathogenic proteins will require high resolution transcriptomics and comparative genomics approaches across soil microbes with different lifestyles. (Summary by Amey Redkar) (Image Courtesy of Pacific Northwest National Laboratory.) Mol. Plant. Pathol. 10.1111/mpp.12628/full
To understand disease development, effector research has mainly focused on the direct interaction of pathogen-derived molecules with plant host targets, or their sensing by surface or intracellular receptors. Recently, attention has turned to the plant microbiome and its key role in maintaining plant health. How microbial communities are manipulated by plant pathogens in sensing and infection of hosts is still largely unexplored. In this Opinion article, Snelders et al. hypothesize the involvement of highly specialized effectors that may act as tools for interaction with beneficial microbes. These microbiome-manipulating effectors may have different transcriptional profiles from other effectors, due to being involved in processes before the actual host contact. Therefore, identifying this interesting set of pathogenic proteins will require high resolution transcriptomics and comparative genomics approaches across soil microbes with different lifestyles. (Summary by Amey Redkar) (Image Courtesy of Pacific Northwest National Laboratory.) Mol. Plant. Pathol. 10.1111/mpp.12628/full
Review. Rhizobia: From saprophytes to endosymbionts (Nat. Rev. Microbiol.) ($)
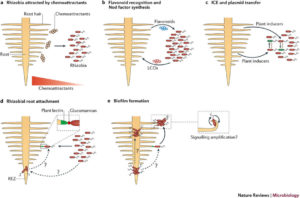 One of the best characterized plant-bacteria interactions is that between legumes and rhizobia. This review by Poole et al. explores rhizobia in their non-plant associated state (as saprophytes that derive energy and nutrients from organic matter in the soil), through the complex signals that lead to their attraction and attachment to the root, and finally through their developmental transition to endosymbiotic bacteroids. The authors observe that many studies of rhizobia focus on the bacteria in pure culture conditions, making it difficult to assess how their interactions with other species in the rhizosphere affects their transition from saprophyte to endosymbiont; these interactions are highlighted in this review. (Summary by Mary Williams) Nat. Rev. Microbiol. 10.1038/nrmicro.2017.171
One of the best characterized plant-bacteria interactions is that between legumes and rhizobia. This review by Poole et al. explores rhizobia in their non-plant associated state (as saprophytes that derive energy and nutrients from organic matter in the soil), through the complex signals that lead to their attraction and attachment to the root, and finally through their developmental transition to endosymbiotic bacteroids. The authors observe that many studies of rhizobia focus on the bacteria in pure culture conditions, making it difficult to assess how their interactions with other species in the rhizosphere affects their transition from saprophyte to endosymbiont; these interactions are highlighted in this review. (Summary by Mary Williams) Nat. Rev. Microbiol. 10.1038/nrmicro.2017.171
Single Parent Expression (SPE) of non-syntenic genes in maize hybrids (Curr. Biol.) ($)
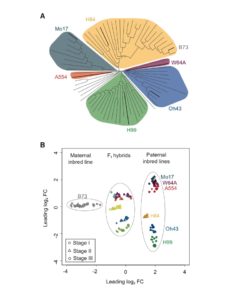 In maize, it has long been known that the crossing of two inbred lines can produce a hybrid offspring with higher yield than the parents. Baldauf and collaborators have studied the gene expression of 6 hybrid lines coming from 7 distantly related inbred lines. One line, B73, was chosen as the common female parent, since it is widely used in breeding programs and has been sequenced. Through RNA sequencing they found that in all hybrids Single Parent Expression (SPE) occurred and that the hybrids expressed between 500 to 600 more genes than the parents. The proportion of non-syntenic genes displaying SPE was also determined; they were overrepresented when compared to all expressed genes. Further analysis of the non-syntenic genes displaying SPE showed that the bZIP and bHLH families of transcription factors were overrepresented, which could explain to a certain degree explain the better performance and plasticity of hybrid maize. (Summary by Cecilia Vasquez-Robinet) Curr. Biol. 10.1016/j.cub.2017.12.027
In maize, it has long been known that the crossing of two inbred lines can produce a hybrid offspring with higher yield than the parents. Baldauf and collaborators have studied the gene expression of 6 hybrid lines coming from 7 distantly related inbred lines. One line, B73, was chosen as the common female parent, since it is widely used in breeding programs and has been sequenced. Through RNA sequencing they found that in all hybrids Single Parent Expression (SPE) occurred and that the hybrids expressed between 500 to 600 more genes than the parents. The proportion of non-syntenic genes displaying SPE was also determined; they were overrepresented when compared to all expressed genes. Further analysis of the non-syntenic genes displaying SPE showed that the bZIP and bHLH families of transcription factors were overrepresented, which could explain to a certain degree explain the better performance and plasticity of hybrid maize. (Summary by Cecilia Vasquez-Robinet) Curr. Biol. 10.1016/j.cub.2017.12.027
Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED 1 (Plant Physiol.)
 Communication between chloroplast and the nucleus is crucial to accomodate changes in the environment as well as regulate development of the chloroplast itself. Five GENOMES UNCOUPLED (GUN2 to -6) genes were previously described to regulate plastid-to-nucleus communication by affecting the synthesis of tetrapyrrole, constituting the active core of chlorophyll. GUN1 does not affect tetrapyrrole biosynthesis. The locus encodes a petatricopeptide repeat protein (PPR) that is involved in protein-to-protein interactions and chloroplast differentiation. GUN1 has a very short lifespan, as is it constantly degraded by Clp protease, with PPR motifs playing a key role in GUN1 protein stability. Although lines overexpressing GUN1 exhibited early flowering phenotypes, the involvement of retrograde signaling in transition from vegetative to reproductive stage requires further examination. (Summary by Magdalena Julkowska) Plant Physiol. 10.1104/pp.18.00009
Communication between chloroplast and the nucleus is crucial to accomodate changes in the environment as well as regulate development of the chloroplast itself. Five GENOMES UNCOUPLED (GUN2 to -6) genes were previously described to regulate plastid-to-nucleus communication by affecting the synthesis of tetrapyrrole, constituting the active core of chlorophyll. GUN1 does not affect tetrapyrrole biosynthesis. The locus encodes a petatricopeptide repeat protein (PPR) that is involved in protein-to-protein interactions and chloroplast differentiation. GUN1 has a very short lifespan, as is it constantly degraded by Clp protease, with PPR motifs playing a key role in GUN1 protein stability. Although lines overexpressing GUN1 exhibited early flowering phenotypes, the involvement of retrograde signaling in transition from vegetative to reproductive stage requires further examination. (Summary by Magdalena Julkowska) Plant Physiol. 10.1104/pp.18.00009
Asymmetric auxin distribution and root tropisms (Plant Cell Physiol.) ($)
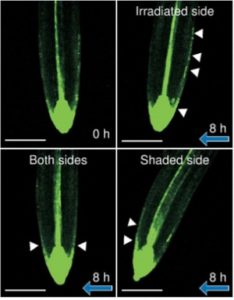 The asymmetric distribution of the phytohormone auxin is essential for tropic responses of seed plants but it is unclear if this distribution is essential for root negative phototropism as well. To understand the role of auxin distribution in root negative phototropism in Arabidopsis, Kimura and colleagues used pharmacological and genetic analyses of wildtype, auxin biosynthesis mutants and auxin receptor mutants to show that polar auxin transport and auxin biosynthesis are critical for root gravitropism but not root phototropism. However, TIR1/AFB auxin receptors and ARF transcriptional factors do play a minor role in root phototropism. Nevertheless, this paper describes that phototropic responses are induced by a process that doesn’t require asymmetric auxin distribution in the roots of Arabidopsis. (Summary by Julia Miller) (Plant Cell Physiol.) 10.1093/pcp/pcy018
The asymmetric distribution of the phytohormone auxin is essential for tropic responses of seed plants but it is unclear if this distribution is essential for root negative phototropism as well. To understand the role of auxin distribution in root negative phototropism in Arabidopsis, Kimura and colleagues used pharmacological and genetic analyses of wildtype, auxin biosynthesis mutants and auxin receptor mutants to show that polar auxin transport and auxin biosynthesis are critical for root gravitropism but not root phototropism. However, TIR1/AFB auxin receptors and ARF transcriptional factors do play a minor role in root phototropism. Nevertheless, this paper describes that phototropic responses are induced by a process that doesn’t require asymmetric auxin distribution in the roots of Arabidopsis. (Summary by Julia Miller) (Plant Cell Physiol.) 10.1093/pcp/pcy018
Knock-down of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development (Plant Physiol.)
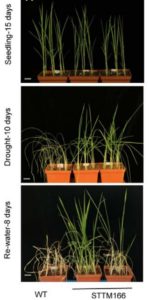 Under drought conditions, moderate leaf rolling improves yield in rice by reducing water loss and allowing efficient photosynthesis (leaf upright position). In STTM166 transgenic rice plants, miRNA166 is knocked down (STTM means “short tandem target mimic”) and the plants constitutively exhibit the rolled-leaf trait. Compared to wild-type plants, STTM166 plants exhibit an improved ability to grow new leaves after a period of water deficit, an increase in spikelet fertility under drought conditions and a decrease in leaf transpiration—all of which contribute to drought tolerance. The changes observed are ABA-independent and are due to modifications in leaf morphology. In addition, STTM166 plants have narrower xylem vessels, resulting in decreased shoot hydraulic conductivity compared to wild-type plants. The target of miRNA166 responsible for rolled-leaf trait is OsHB4, a homeodomain leucine zipper III transcription factor involved in adaxial polarity of the leaf. Rice plants overexpressing OsBH4 have improved drought tolerance. Through transcriptomic analysis, polysaccharide synthase genes are shown to be the downstream targets of the miR166-OsBH4 regulatory module. This study gives evidence to the importance of regulating leaf morphology to increase drought tolerance in rice. (Summary by Stephanie Saade) Plant Physiol. 10.1104/pp.17.01432
Under drought conditions, moderate leaf rolling improves yield in rice by reducing water loss and allowing efficient photosynthesis (leaf upright position). In STTM166 transgenic rice plants, miRNA166 is knocked down (STTM means “short tandem target mimic”) and the plants constitutively exhibit the rolled-leaf trait. Compared to wild-type plants, STTM166 plants exhibit an improved ability to grow new leaves after a period of water deficit, an increase in spikelet fertility under drought conditions and a decrease in leaf transpiration—all of which contribute to drought tolerance. The changes observed are ABA-independent and are due to modifications in leaf morphology. In addition, STTM166 plants have narrower xylem vessels, resulting in decreased shoot hydraulic conductivity compared to wild-type plants. The target of miRNA166 responsible for rolled-leaf trait is OsHB4, a homeodomain leucine zipper III transcription factor involved in adaxial polarity of the leaf. Rice plants overexpressing OsBH4 have improved drought tolerance. Through transcriptomic analysis, polysaccharide synthase genes are shown to be the downstream targets of the miR166-OsBH4 regulatory module. This study gives evidence to the importance of regulating leaf morphology to increase drought tolerance in rice. (Summary by Stephanie Saade) Plant Physiol. 10.1104/pp.17.01432
A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi (PNAS) ($)
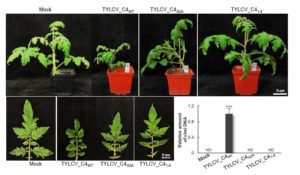 Viruses can move from cell to cell through plasmodesmata. Small interfering RNAs (siRNAs) are key components in the plant’s arsenal against viruses. They function by harnessing the AGO system to target and cleave viral RNA, thus silencing the viruses. Like the viruses they target, siRNAs move from cell to cell through plasmodesmata. Previous work showed that the C4 protein from the Tomato yellow leaf curl virus (TYLCV) can delay the spread of viral silencing. Through some cleverly designed experiments, Rosas-Diaz et al. showed that a previously identified plant protein BAM1 (BARELY ANY MERISTEM 1) binds to the viral C4 protein, and contributes to the spread of siRNAs. The authors propose that the function of the viral C4 protein is to support viral pathogenicity by interfering with the spread of silencing siRNAs through an interaction with BAM1. As BAM1 also has a role in development, it will be interesting to determine if its developmental function is also mediated by interaction with small RNAs, which are known to contribute to developmental patterning. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1715556115
Viruses can move from cell to cell through plasmodesmata. Small interfering RNAs (siRNAs) are key components in the plant’s arsenal against viruses. They function by harnessing the AGO system to target and cleave viral RNA, thus silencing the viruses. Like the viruses they target, siRNAs move from cell to cell through plasmodesmata. Previous work showed that the C4 protein from the Tomato yellow leaf curl virus (TYLCV) can delay the spread of viral silencing. Through some cleverly designed experiments, Rosas-Diaz et al. showed that a previously identified plant protein BAM1 (BARELY ANY MERISTEM 1) binds to the viral C4 protein, and contributes to the spread of siRNAs. The authors propose that the function of the viral C4 protein is to support viral pathogenicity by interfering with the spread of silencing siRNAs through an interaction with BAM1. As BAM1 also has a role in development, it will be interesting to determine if its developmental function is also mediated by interaction with small RNAs, which are known to contribute to developmental patterning. (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1715556115
Auxin synthesis contributes to virulence of Pseudomonas syringae (PLOS Pathogens)
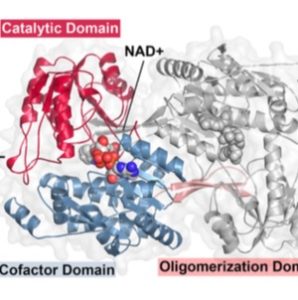 Plant pathogens have developed a large range of strategies to allow them to have successful interactions with their plant host, including physiological manipulation. For example, Pseudomonas syringae, the cause of speck disease in many plant systems, manipulates the auxin phytohormone physiology in its plant hosts. McClerklin et al. showed that P. syringae pv. tomato strain DC3000 synthesizes indole-3-acetic acid (IAA) via an indole-3-acetaldehyde intermediate. The researchers also identified the putative genes of PstDC3000 aldehyde dehydrogenase and performed a biochemical analysis of their corresponding protein, including the overall three-dimensional structure of indole-3-acetaldehyde dehydrogenase (AldA) and the structure of its active site. The use of DC3000 IAA biosynthesis mutants (ald mutants), which produce less IAA in culture in comparison with the DC3000 controls, showed that ald mutants exhibit reduced virulence on Arabidopsis thaliana, demonstrating that auxin contributes to virulence in P. syringae. Furthermore, the authors showed that pathogen-derived IAA suppresses salicylic acid-mediated defenses, while plant-derived IAA promotes pathogen growth, showing that IAA plays multiple roles during pathogenesis through different mechanisms depending on whether it originates in plant or pathogen. (Summary by Isabel Mendoza) PLOS Pathogens 10.1371/journal.ppat.1006811
Plant pathogens have developed a large range of strategies to allow them to have successful interactions with their plant host, including physiological manipulation. For example, Pseudomonas syringae, the cause of speck disease in many plant systems, manipulates the auxin phytohormone physiology in its plant hosts. McClerklin et al. showed that P. syringae pv. tomato strain DC3000 synthesizes indole-3-acetic acid (IAA) via an indole-3-acetaldehyde intermediate. The researchers also identified the putative genes of PstDC3000 aldehyde dehydrogenase and performed a biochemical analysis of their corresponding protein, including the overall three-dimensional structure of indole-3-acetaldehyde dehydrogenase (AldA) and the structure of its active site. The use of DC3000 IAA biosynthesis mutants (ald mutants), which produce less IAA in culture in comparison with the DC3000 controls, showed that ald mutants exhibit reduced virulence on Arabidopsis thaliana, demonstrating that auxin contributes to virulence in P. syringae. Furthermore, the authors showed that pathogen-derived IAA suppresses salicylic acid-mediated defenses, while plant-derived IAA promotes pathogen growth, showing that IAA plays multiple roles during pathogenesis through different mechanisms depending on whether it originates in plant or pathogen. (Summary by Isabel Mendoza) PLOS Pathogens 10.1371/journal.ppat.1006811
The biotrophic development of Ustilago maydis studied by RNAseq analysis (Plant Cell)
 The corn smut fungus Ustilago maydis that causes tumorous symptoms on all aerial parts of maize has established itself as a model system to dissect host colonization strategies by biotrophic fungi. Transcriptional responses upon U. maydis colonization were previously demonstrated by several studies using an engineered solopathogenic strain of U. maydis. However, a time-resolved transcriptional profile across different plant-associated stages in U. maydis lifecycle was still unknown. Lanver et al. have filled this gap by performing an in depth transcriptional profiling across different plant-associated developmental stages using compatible haploid wild-type strains. The authors focused on genes involved in fungal metabolism, nutritional strategies, secreted effectors and regulatory networks. Waves of effector expression were regulated across different infection stages during symptom formation. Several nutrient transporters were linked to effector expression, showing nutrient dependent as well as independent expression. This work also identified several yet uncharacterized transcription factors that likely play a role in tumour development. Hence this study provides a new resource to understand complex symptom formation and modulation and acquisition of nutrients by fungi in the plant environment. (Summary by Amey Redkar) Plant. Cell. 10.1105/tpc.17.00764
The corn smut fungus Ustilago maydis that causes tumorous symptoms on all aerial parts of maize has established itself as a model system to dissect host colonization strategies by biotrophic fungi. Transcriptional responses upon U. maydis colonization were previously demonstrated by several studies using an engineered solopathogenic strain of U. maydis. However, a time-resolved transcriptional profile across different plant-associated stages in U. maydis lifecycle was still unknown. Lanver et al. have filled this gap by performing an in depth transcriptional profiling across different plant-associated developmental stages using compatible haploid wild-type strains. The authors focused on genes involved in fungal metabolism, nutritional strategies, secreted effectors and regulatory networks. Waves of effector expression were regulated across different infection stages during symptom formation. Several nutrient transporters were linked to effector expression, showing nutrient dependent as well as independent expression. This work also identified several yet uncharacterized transcription factors that likely play a role in tumour development. Hence this study provides a new resource to understand complex symptom formation and modulation and acquisition of nutrients by fungi in the plant environment. (Summary by Amey Redkar) Plant. Cell. 10.1105/tpc.17.00764
Calling from distance: attraction of soil bacteria by plant root volatiles (ISME J.) ($)
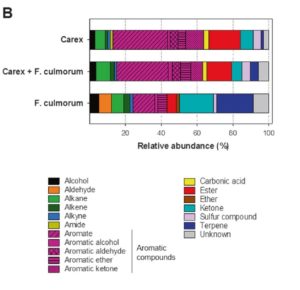 Plant interactions with beneficial microbes are good strategies to survive biotic and abiotic challenging conditions, but the mechanisms that plants use to recruit these interactions, specially belowground, remain scarcely known. Schulz-Bohm et al. designed an olfactometer system to analyze the long distance interactions between soil bacteria and volatile organic compounds (VOCs) emitted by Carex arenaria roots. They also introduced the fungal pathogen Fusarium culmorum, to see if the presence of the pathogen affected the plants’ VOCs emissions and the presence of bacteria. Interestingly, their results showed that plant VOCs can attract a specific set of soil bacteria from long distance, as they can provide nutrient-environment information, and the microbes can differentiate between a rich- or poor- nutrient environment. In addition, when challenged with a fungal pathogen, the root VOCs changed and bacteria with antifungal properties were attracted. This olfactometer system, which resembles more closely the natural conditions belowground, can easily be used for further studies. (Summary by Maria Julissa Ek-Ramos) ISME Journal. 10.1038/s41396-017-0035-3
Plant interactions with beneficial microbes are good strategies to survive biotic and abiotic challenging conditions, but the mechanisms that plants use to recruit these interactions, specially belowground, remain scarcely known. Schulz-Bohm et al. designed an olfactometer system to analyze the long distance interactions between soil bacteria and volatile organic compounds (VOCs) emitted by Carex arenaria roots. They also introduced the fungal pathogen Fusarium culmorum, to see if the presence of the pathogen affected the plants’ VOCs emissions and the presence of bacteria. Interestingly, their results showed that plant VOCs can attract a specific set of soil bacteria from long distance, as they can provide nutrient-environment information, and the microbes can differentiate between a rich- or poor- nutrient environment. In addition, when challenged with a fungal pathogen, the root VOCs changed and bacteria with antifungal properties were attracted. This olfactometer system, which resembles more closely the natural conditions belowground, can easily be used for further studies. (Summary by Maria Julissa Ek-Ramos) ISME Journal. 10.1038/s41396-017-0035-3
A mucin-like protein of planthopper is required for feeding and induces immunity response in plants (Plant Physiol.)
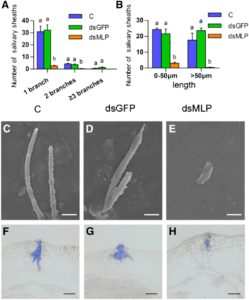 Piercing-sucking insects secrete saliva while feeding through specialized mouth parts called stylets. Saliva plays critical roles in insect digestion and nutrition, and regulates plant defense responses, but the specific metabolites involved have not been characterized. Shangguan et al. reported the identification of a specific protein secreted in the saliva of Nilaparvata lugens, an important rice pest. This protein is a mucin-like protein (NIMLP), highly expressed in salivary glands and secreted to rice while feeding. Expression of this gene was necessary for the formation of salivary sheaths in the insect as demonstrated by silencing, which affected insect development and ultimately reduced salivary sheath formation, feeding, and performance. In addition, in the plant NIMLP induced callose deposition, expression of defense-related genes (by Ca2+ mobilization and MEK2 MAP kinase and jasmonic acid signaling pathways) and induction of cell death. Detailed experiments also showed that the functional portion of NIMLP is located at the C-terminus and that the plant receptor that recognizes NIMLP remains to be identified. (Summary by Maria Julissa Ek-Ramos) Plant Physiol. 10.1104/pp.17.00755
Piercing-sucking insects secrete saliva while feeding through specialized mouth parts called stylets. Saliva plays critical roles in insect digestion and nutrition, and regulates plant defense responses, but the specific metabolites involved have not been characterized. Shangguan et al. reported the identification of a specific protein secreted in the saliva of Nilaparvata lugens, an important rice pest. This protein is a mucin-like protein (NIMLP), highly expressed in salivary glands and secreted to rice while feeding. Expression of this gene was necessary for the formation of salivary sheaths in the insect as demonstrated by silencing, which affected insect development and ultimately reduced salivary sheath formation, feeding, and performance. In addition, in the plant NIMLP induced callose deposition, expression of defense-related genes (by Ca2+ mobilization and MEK2 MAP kinase and jasmonic acid signaling pathways) and induction of cell death. Detailed experiments also showed that the functional portion of NIMLP is located at the C-terminus and that the plant receptor that recognizes NIMLP remains to be identified. (Summary by Maria Julissa Ek-Ramos) Plant Physiol. 10.1104/pp.17.00755