What We’re Reading: December 22nd
Review: On the selectivity, specificity and signaling potential of long-distance movement of messenger RNA
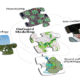
 Regulation of transcription occurs at the cell-type specific level, but transcribed messenger RNA is mobile and can move between tissues through plant vasculature, serving as a long distance messenger. Many mRNA molecules have been identified in the phloem sap, suggesting that mRNA transport goes through plasmodesmata from companion cells into the phloem sieve elements. Transport of mRNA encoding a broad range of functions occurs at the genomic scale but at low efficiency. The secondary structure motifs in the 3′UTR seem to be sufficient for mRNA transport, and interaction with RNA-binding proteins might enhance transcript stability and transport. In the future, visualization of individual transcripts and determining the interactions between mRNA and binding proteins will shed more light on the mechanisms underlying long-distance mRNA transport. (Summary by Magdalena Julkowska) Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.11.001
Regulation of transcription occurs at the cell-type specific level, but transcribed messenger RNA is mobile and can move between tissues through plant vasculature, serving as a long distance messenger. Many mRNA molecules have been identified in the phloem sap, suggesting that mRNA transport goes through plasmodesmata from companion cells into the phloem sieve elements. Transport of mRNA encoding a broad range of functions occurs at the genomic scale but at low efficiency. The secondary structure motifs in the 3′UTR seem to be sufficient for mRNA transport, and interaction with RNA-binding proteins might enhance transcript stability and transport. In the future, visualization of individual transcripts and determining the interactions between mRNA and binding proteins will shed more light on the mechanisms underlying long-distance mRNA transport. (Summary by Magdalena Julkowska) Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.11.001
Review. Grasses: The original Vikings ($)
 The Vikings were notorious raiders for centuries, pillaging and looting the shores throughout the northern hemisphere. Through their successful raids, the Vikings established colonies that grew into states and countries, among these Normandy, England, Sicily, and Russia. The success of the Vikings is due to several factors, including effective dispersal, rapid population growth, resilience, plasticity, and transforming their environments to fit their needs. A recent review by Linder et al. discusses how “Viking Syndrome” (invasive ability) is responsible for the success of the grasses (Poaceae), allowing the most successful angiosperm family to establish and thrive around the globe. The grasses developed key attributes at the right times in their evolutionary history. The dispersal ability of the grasses allowed them to reach nearly all four corners of the Earth, and is largely made possible by the spikelet, an inflorescence structure unique to them. Grasses are also highly successful at establishing in new environments due to their short generation times, and capable of surviving genetic bottlenecks by wind pollination, occurrence of polyploidy, and self-incompatibility. C3 and C4 photosynthesis, frost tolerance, and flexible growth forms allow grasses to dominate a range of environmental extremes, and a high tolerance of environmental disturbance, such as fire and herbivores, allow grasses to not only persist, but thrive in environments that limit the growth of most other plants. Grasses, having become incredibly successful over the last 100 million years, did not truly attain their success through the Viking Syndrome, rather, the Vikings triumphed because of the Grass Syndrome. (Summary by Danielle Roodt Prinsloo) Biol. Rev. 10.1111/brv.12388
The Vikings were notorious raiders for centuries, pillaging and looting the shores throughout the northern hemisphere. Through their successful raids, the Vikings established colonies that grew into states and countries, among these Normandy, England, Sicily, and Russia. The success of the Vikings is due to several factors, including effective dispersal, rapid population growth, resilience, plasticity, and transforming their environments to fit their needs. A recent review by Linder et al. discusses how “Viking Syndrome” (invasive ability) is responsible for the success of the grasses (Poaceae), allowing the most successful angiosperm family to establish and thrive around the globe. The grasses developed key attributes at the right times in their evolutionary history. The dispersal ability of the grasses allowed them to reach nearly all four corners of the Earth, and is largely made possible by the spikelet, an inflorescence structure unique to them. Grasses are also highly successful at establishing in new environments due to their short generation times, and capable of surviving genetic bottlenecks by wind pollination, occurrence of polyploidy, and self-incompatibility. C3 and C4 photosynthesis, frost tolerance, and flexible growth forms allow grasses to dominate a range of environmental extremes, and a high tolerance of environmental disturbance, such as fire and herbivores, allow grasses to not only persist, but thrive in environments that limit the growth of most other plants. Grasses, having become incredibly successful over the last 100 million years, did not truly attain their success through the Viking Syndrome, rather, the Vikings triumphed because of the Grass Syndrome. (Summary by Danielle Roodt Prinsloo) Biol. Rev. 10.1111/brv.12388
LTR_retriever: a highly accurate and sensitive program for identification of long terminal-repeat retrotransposons
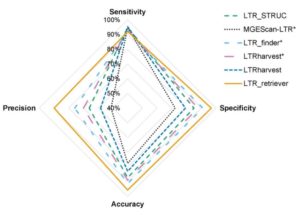 Transposable elements are a major part of plant genomes. Long-terminal repeat retrotransposons (LTR (LTR-RTs) alone make up 78% of the maize genome (retrotransposons use a “copy and paste” transposition method meaning that a single source element can generate numerous clones of itself, leading to tremendous amplification). In the absence of selection, LTR-RTs are particularly subject to mutation, meaning that there is considerable variation in their structure and sequence. Therefore, efforts to sequence and annotate genomes will be greatly enhanced by the ability to reliably and sensitively recognize LTR-RTs. Ou and Jiang have developed a program, LTR_retriever, to annotate LTR-RTs in plant genomes, and demonstrated its effectiveness in several monocot and dicot genomes. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.01310
Transposable elements are a major part of plant genomes. Long-terminal repeat retrotransposons (LTR (LTR-RTs) alone make up 78% of the maize genome (retrotransposons use a “copy and paste” transposition method meaning that a single source element can generate numerous clones of itself, leading to tremendous amplification). In the absence of selection, LTR-RTs are particularly subject to mutation, meaning that there is considerable variation in their structure and sequence. Therefore, efforts to sequence and annotate genomes will be greatly enhanced by the ability to reliably and sensitively recognize LTR-RTs. Ou and Jiang have developed a program, LTR_retriever, to annotate LTR-RTs in plant genomes, and demonstrated its effectiveness in several monocot and dicot genomes. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.01310
Methylome robustness in plants is conferred by a methylation-sensitive system.
 DNA methylation is instrumental in promoting transcriptional silencing at repetitive elements, inhibiting illegitimate recombination and establishing genomic imprinting. In plants, DNA methylation profiles are stably inherited over generations through the activities of cytosine DNA methyltransferases and 5-methylcytosine glycosylases. Nevertheless, how they interact and influence each other’s activities is not well understood. In this study, Williams and Gehring identify a methylation-sensing gene regulatory system centered on the DNA glycosylase ROS1. ROS1 expression has been described previously as being directly regulated by DNA methylation, and as a consequence it is down-regulated in methylation mutants. By engineering Arabidopsis plants expressing a version of ROS1 insensitive to methylation loss, the authors show that expression of ROS1 in a methylation mutant causes aberrant phenotypes that are amplified over generations. Whole-genome bisulfite sequencing revealed widespread demethylation across the genome. The changes in methylation were not stable through successive generations, with heterochromatin progressively regaining methylation in stark contrast with loci located in euchromatin showing cumulative loss of methylation over generations. Altogether, this study shows that methylation and demethylation pathways are intimately linked within a methylation-sensing system that buffers instability and variations of the methylome. It further supports the idea that the epigenome is particularly robust to destabilizing stimuli. (Summary by Matthias Benoit) Nature Comms. 10.1038/s41467-017-02219-3
DNA methylation is instrumental in promoting transcriptional silencing at repetitive elements, inhibiting illegitimate recombination and establishing genomic imprinting. In plants, DNA methylation profiles are stably inherited over generations through the activities of cytosine DNA methyltransferases and 5-methylcytosine glycosylases. Nevertheless, how they interact and influence each other’s activities is not well understood. In this study, Williams and Gehring identify a methylation-sensing gene regulatory system centered on the DNA glycosylase ROS1. ROS1 expression has been described previously as being directly regulated by DNA methylation, and as a consequence it is down-regulated in methylation mutants. By engineering Arabidopsis plants expressing a version of ROS1 insensitive to methylation loss, the authors show that expression of ROS1 in a methylation mutant causes aberrant phenotypes that are amplified over generations. Whole-genome bisulfite sequencing revealed widespread demethylation across the genome. The changes in methylation were not stable through successive generations, with heterochromatin progressively regaining methylation in stark contrast with loci located in euchromatin showing cumulative loss of methylation over generations. Altogether, this study shows that methylation and demethylation pathways are intimately linked within a methylation-sensing system that buffers instability and variations of the methylome. It further supports the idea that the epigenome is particularly robust to destabilizing stimuli. (Summary by Matthias Benoit) Nature Comms. 10.1038/s41467-017-02219-3
The plastid lipocalin LCNP is required for sustained photoprotective energy dissipation
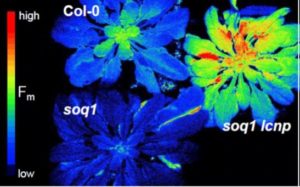 Plants have several mechanisms to protect themselves from damage from excess light, including a set of reactions collectively described as non-photochemical quenching or NPQ. One of these is a sustained and slowly reversible form of NPQ, which the authors have named qH. How this sustained NPQ functions is not known. Starting with a previously identified double mutant, soq1 (suppressor of quenching1) npq4 (non-photochemical quenching4), which has enhanced qH, Malnoë et al. carried out a genetic screen to identify suppressors. One of the mutants they isolated affects chloroplastic lipocalin, newly renamed as LCNP (to avoid confusion with the common abbreviation for chlorophyll, CHL). Lipocalins are small proteins that bind small hydrophobic molecules. Although the mechanism remains unclear, the authors show that LCNP protects the thylakoid membrane and prevents singlet oxygen stress by enabling sustained NPQ in the peripheral antenna (LHCII) of photosystem II. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00536
Plants have several mechanisms to protect themselves from damage from excess light, including a set of reactions collectively described as non-photochemical quenching or NPQ. One of these is a sustained and slowly reversible form of NPQ, which the authors have named qH. How this sustained NPQ functions is not known. Starting with a previously identified double mutant, soq1 (suppressor of quenching1) npq4 (non-photochemical quenching4), which has enhanced qH, Malnoë et al. carried out a genetic screen to identify suppressors. One of the mutants they isolated affects chloroplastic lipocalin, newly renamed as LCNP (to avoid confusion with the common abbreviation for chlorophyll, CHL). Lipocalins are small proteins that bind small hydrophobic molecules. Although the mechanism remains unclear, the authors show that LCNP protects the thylakoid membrane and prevents singlet oxygen stress by enabling sustained NPQ in the peripheral antenna (LHCII) of photosystem II. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00536
Cell density and airspace patterning in the leaf can be manipulated to increase leaf photosynthetic capacity
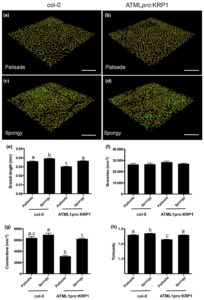 Increasing photosynthetic conversion efficiency is an attractive target for improving crop yields. One way of affecting this is to alter the way CO2 is delivered to Rubisco, the carbon-fixing enzyme of photosynthesis. Lehmeier et al. aimed to change the pattern of air spaces within Arabidopsis leaves by modulating the expression of the cell cycle genes KRP1 and RBR1. They were able to manipulate mesophyll cell size, and used microCT imaging to demonstrate that this affected the architecture of air channels. These manipulations led to an increase in photosynthetic rate, which could only partially be explained by changes in leaf thickness. Increases in Rubisco activity and mesophyll conductance were also observed, indicating that this could be a viable strategy for boosting photosynthetic assimilation. (Summary by Mike Page) Plant J. 10.1111/tpj.13727
Increasing photosynthetic conversion efficiency is an attractive target for improving crop yields. One way of affecting this is to alter the way CO2 is delivered to Rubisco, the carbon-fixing enzyme of photosynthesis. Lehmeier et al. aimed to change the pattern of air spaces within Arabidopsis leaves by modulating the expression of the cell cycle genes KRP1 and RBR1. They were able to manipulate mesophyll cell size, and used microCT imaging to demonstrate that this affected the architecture of air channels. These manipulations led to an increase in photosynthetic rate, which could only partially be explained by changes in leaf thickness. Increases in Rubisco activity and mesophyll conductance were also observed, indicating that this could be a viable strategy for boosting photosynthetic assimilation. (Summary by Mike Page) Plant J. 10.1111/tpj.13727
Role of C-terminal modules in phytochrome signaling
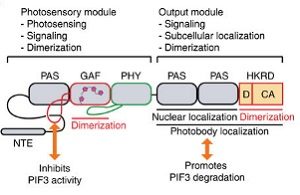 Phytochromes are photoreceptors found in many organisms including bacteria, fungi and plants. Plant phytochromes regulate many aspects of plant growth and development, including germination, shade avoidance, defense against pests, and senescence. Phytochromes most likely transduce signals through the degradation of phytochrome-interacting transcription factors (PIFs) by the N-terminal photosensory module. The C-terminal module, which includes a histidine kinase-related domain (HKRD), is thought to not participate in signaling. However, Qui and colleagues report that the C-terminal module of the Arabidopsis phytochrome B (PHYB) causes the degradation of PIF3 and activates genes related to photosynthesis. This is due to the role of HKRP as a dimerization domain for the homodimers and heterodimers of PHYB. If HKRP is modified there is no longer PIF3 degradation, while modifications of the N-terminal module have little effect on PIF3 degradation. This paper describes the important role of the C-terminal modules of phyotochromes in dimerization and signaling. (Summary by Julia Miller) Nature Comms. 10.1038/s41467-017-02062-6
Phytochromes are photoreceptors found in many organisms including bacteria, fungi and plants. Plant phytochromes regulate many aspects of plant growth and development, including germination, shade avoidance, defense against pests, and senescence. Phytochromes most likely transduce signals through the degradation of phytochrome-interacting transcription factors (PIFs) by the N-terminal photosensory module. The C-terminal module, which includes a histidine kinase-related domain (HKRD), is thought to not participate in signaling. However, Qui and colleagues report that the C-terminal module of the Arabidopsis phytochrome B (PHYB) causes the degradation of PIF3 and activates genes related to photosynthesis. This is due to the role of HKRP as a dimerization domain for the homodimers and heterodimers of PHYB. If HKRP is modified there is no longer PIF3 degradation, while modifications of the N-terminal module have little effect on PIF3 degradation. This paper describes the important role of the C-terminal modules of phyotochromes in dimerization and signaling. (Summary by Julia Miller) Nature Comms. 10.1038/s41467-017-02062-6
Reassessing the evolution of strigolactone synthesis and signaling
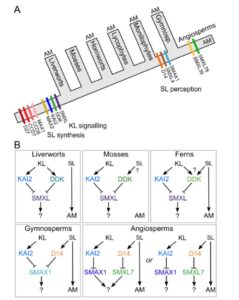 Much of our understanding of strigolactones (SLs) as developmental hormones and rhizosphere signals comes from studies of angiosperms. Understanding the ancestral role for strigolactones is complicated by the fact that some of the SL-related genes are closely related to those responsive to karrikins (smoke-derived factors), and as yet unidentified endogenous karrikin-like (KL) factors. Walker and Bennett have explored numerous genomic and transcriptomic databases spanning the diversity of major plant clades to learn about the evolutionary origins of SL and KL functions. By identifying which biosynthetic, receptor and downstream genes are present in each of the plant lineages, they propose a model for the roles of strigolactones and karrikin-like factors in each of these lineages. Their results suggest that the KL-signaling module is ancestral, and that SLs may have originated as rhizosphere signaling molecules that were relatively recently recruited as internal hormones. (Summary by Mary Williams) bioRxiv 10.1101/228320
Much of our understanding of strigolactones (SLs) as developmental hormones and rhizosphere signals comes from studies of angiosperms. Understanding the ancestral role for strigolactones is complicated by the fact that some of the SL-related genes are closely related to those responsive to karrikins (smoke-derived factors), and as yet unidentified endogenous karrikin-like (KL) factors. Walker and Bennett have explored numerous genomic and transcriptomic databases spanning the diversity of major plant clades to learn about the evolutionary origins of SL and KL functions. By identifying which biosynthetic, receptor and downstream genes are present in each of the plant lineages, they propose a model for the roles of strigolactones and karrikin-like factors in each of these lineages. Their results suggest that the KL-signaling module is ancestral, and that SLs may have originated as rhizosphere signaling molecules that were relatively recently recruited as internal hormones. (Summary by Mary Williams) bioRxiv 10.1101/228320
Microtubule involvement in stomatal dynamics
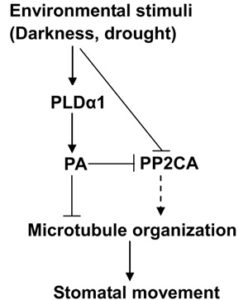 Stomatal dynamics have been investigated in terms of ion exchange involvements, turgor pressure fluctuations, and transcriptional changes, particularly in response to environmental stresses. However, the involvement of the cytoskeleton during this process, particularly concerning the role of microtubules (MTs), is a relatively new field of study. Qu et al. investigated the role of MTs in ABA- and light-induced stomatal responses. Stabilizing MTs with paclitaxel promoted light-induced stomatal opening, with guard cells exhibiting more organized and denser MT arrays, but this MT stabilization inhibited dark-induced closure. On the other hand, destabilizing MTs inhibited light-induced opening and abolished the MT network but did not affect dark-induced closure. ABA does disrupt MTs and the authors found that paclitaxel partially arrested ABA-induced closure and MT depolymerization. The ABA-insensitive pldα1 mutants were able to be rescued with cotreatment of ABA and oryzalin, but not oryzalin alone. These data suggest that MT reorganization is required but not sufficient for ABA-induced stomatal closure and that MTs are involved in stomatal opening. (Summary by Alecia Biel) Plant Growth Regulation 10.1007/s10725-017-0353-5.
Stomatal dynamics have been investigated in terms of ion exchange involvements, turgor pressure fluctuations, and transcriptional changes, particularly in response to environmental stresses. However, the involvement of the cytoskeleton during this process, particularly concerning the role of microtubules (MTs), is a relatively new field of study. Qu et al. investigated the role of MTs in ABA- and light-induced stomatal responses. Stabilizing MTs with paclitaxel promoted light-induced stomatal opening, with guard cells exhibiting more organized and denser MT arrays, but this MT stabilization inhibited dark-induced closure. On the other hand, destabilizing MTs inhibited light-induced opening and abolished the MT network but did not affect dark-induced closure. ABA does disrupt MTs and the authors found that paclitaxel partially arrested ABA-induced closure and MT depolymerization. The ABA-insensitive pldα1 mutants were able to be rescued with cotreatment of ABA and oryzalin, but not oryzalin alone. These data suggest that MT reorganization is required but not sufficient for ABA-induced stomatal closure and that MTs are involved in stomatal opening. (Summary by Alecia Biel) Plant Growth Regulation 10.1007/s10725-017-0353-5.
DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in rice
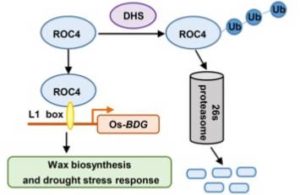 Wax covering the outer surface of the shoot (epicuticular wax) is crucial in the ability of the plant to conserve water. Wang et al. identified a drought hypersensitive plant that overexpresses an E3 ubiquitin ligase which they named DROUGHT HYPERSENSITIVE. In these overexpression plants, there was a significant reduction in wax deposition. The authors speculated that DHS might function through targeting a transcription factor involved in wax deposition. They screened several known transcription factors for interaction with DHS and found that ROC4, (which has previously been shown to regulate wax biosynthesis in Arabidopsis, maize and tomato) interacts with DHS in vivo and in vitro. Finally, they showed that ROC4 binds to and regulates the expression of BDG, a gene involved in wax biosynthesis. This signaling pathway offers a new target for engineering drought-tolerance. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00823
Wax covering the outer surface of the shoot (epicuticular wax) is crucial in the ability of the plant to conserve water. Wang et al. identified a drought hypersensitive plant that overexpresses an E3 ubiquitin ligase which they named DROUGHT HYPERSENSITIVE. In these overexpression plants, there was a significant reduction in wax deposition. The authors speculated that DHS might function through targeting a transcription factor involved in wax deposition. They screened several known transcription factors for interaction with DHS and found that ROC4, (which has previously been shown to regulate wax biosynthesis in Arabidopsis, maize and tomato) interacts with DHS in vivo and in vitro. Finally, they showed that ROC4 binds to and regulates the expression of BDG, a gene involved in wax biosynthesis. This signaling pathway offers a new target for engineering drought-tolerance. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00823
Extremely flexible infection programs in a fungal plant pathogen
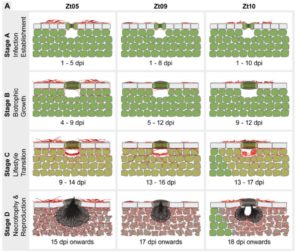 Filamentous plant pathogens have developed an extra-ordinary level of dynamic genome architecture that adapts to changing host environment in the best possible way to promote infection. There are very limited studies describing the impact of this genome plasticity on phenotypic variation. Haueisen et al. have shown the effect of genome variability on infection phenotypes in the hemibiotrophic pathogen of wheat Zymoseptoria tritici. The authors have studied three isolates of this pathogen which were collected across varied geographical regions and which show a variation in genome and gene content. All the isolates cause similar phenotypes on a susceptible wheat cultivar despite the genomic variation. Comparative transcriptomics of these three pathogen isolates across different stages of disease cycle show 20% of the differentially expressed genes (DEGs) between the isolates, a significant number of which encode for effectors. Furthermore, these DEGs fall within the region of transposable elements and show signatures of histone modification. Hence, this study highlights the importance of genome plasticity in fungal plant pathogens that exhibit differential infection programs to adapt to the changing host environment across various geographical regions. (Summary by Amey Redkar) bioRxiv
Filamentous plant pathogens have developed an extra-ordinary level of dynamic genome architecture that adapts to changing host environment in the best possible way to promote infection. There are very limited studies describing the impact of this genome plasticity on phenotypic variation. Haueisen et al. have shown the effect of genome variability on infection phenotypes in the hemibiotrophic pathogen of wheat Zymoseptoria tritici. The authors have studied three isolates of this pathogen which were collected across varied geographical regions and which show a variation in genome and gene content. All the isolates cause similar phenotypes on a susceptible wheat cultivar despite the genomic variation. Comparative transcriptomics of these three pathogen isolates across different stages of disease cycle show 20% of the differentially expressed genes (DEGs) between the isolates, a significant number of which encode for effectors. Furthermore, these DEGs fall within the region of transposable elements and show signatures of histone modification. Hence, this study highlights the importance of genome plasticity in fungal plant pathogens that exhibit differential infection programs to adapt to the changing host environment across various geographical regions. (Summary by Amey Redkar) bioRxiv
Evolutionary transitions between beneficial and phytopathogenic Rhodococcus
 Rodococcus bacteria are often identified as causal agents in disease outbreaks. Savory et al. analyzed 60 isolates from diseased plants. By comparing these new isolates and previous isolates, they found that 64 of 66 pathogenic isolates carry a linear virulence plasmid, and that all but four carried genes previously associated with pathogenicity. A comparison of genomic and plasmid sequences indicated that the pathogenic isolates are more diverse than the plasmid, indicating that the virulence plasmid can be introduced through horizontal gene transfer and can be effective in diverse genetic backgrounds. When they lack the virulence plasmid, many of these isolates are in fact beneficial to plant growth. Therefore, the beneficial microbe can quickly switch to a pathogenic one simply by acquisition of the virulence plasmid. The authors observe that these bacteria are “model organisms for characterizing evolutionary transitions between alternative symbiotic states.” (Summary by Mary Williams) eLIFE 10.7554/eLife.30925
Rodococcus bacteria are often identified as causal agents in disease outbreaks. Savory et al. analyzed 60 isolates from diseased plants. By comparing these new isolates and previous isolates, they found that 64 of 66 pathogenic isolates carry a linear virulence plasmid, and that all but four carried genes previously associated with pathogenicity. A comparison of genomic and plasmid sequences indicated that the pathogenic isolates are more diverse than the plasmid, indicating that the virulence plasmid can be introduced through horizontal gene transfer and can be effective in diverse genetic backgrounds. When they lack the virulence plasmid, many of these isolates are in fact beneficial to plant growth. Therefore, the beneficial microbe can quickly switch to a pathogenic one simply by acquisition of the virulence plasmid. The authors observe that these bacteria are “model organisms for characterizing evolutionary transitions between alternative symbiotic states.” (Summary by Mary Williams) eLIFE 10.7554/eLife.30925