What We’re Reading: August 11
J. Exp. Bot. reviews auxin ($)
 The Journal of Experimental Botany is publishing a good collection of review articles on auxin. Topics include ARF transcription factors, Auxin’s role in lateral root formation, Auxin research in rice and implications for crop improvement, Integration of multiple auxin signaling pathways, Evolution of auxin signaling, Auxin homeostasis, and Auxin, microtubules, and vesicle trafficking. As you know, auxin apparently is involved in everything, so it’s always a good time to refresh your understanding about this important hormone. J. Exp. Bot.
The Journal of Experimental Botany is publishing a good collection of review articles on auxin. Topics include ARF transcription factors, Auxin’s role in lateral root formation, Auxin research in rice and implications for crop improvement, Integration of multiple auxin signaling pathways, Evolution of auxin signaling, Auxin homeostasis, and Auxin, microtubules, and vesicle trafficking. As you know, auxin apparently is involved in everything, so it’s always a good time to refresh your understanding about this important hormone. J. Exp. Bot.
Reviews: Membrane and vesicle trafficking in plant immunity and beyond ($)
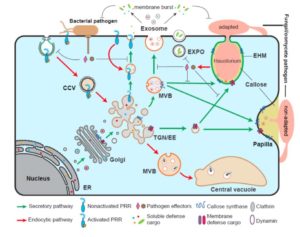 New reviews cover the hot topic of membrane and vesicle trafficking in plant immunity. Gu et al. (Mol. Plant 10.1016/j.molp.2017.07.001) provide an overview of the two membrane trafficking pathways: the secretory pathway involved in movement of antimicrobials, defense proteins and cell wall components outwards, and the endocytic pathway involved in movement of immune receptors inward for signal transduction. They describe the molecular players and regulators, features of these pathways that are unique to plants, how these pathways are involved in defense, and how pathogens sometimes co-opt them to supress immunity. Yun and Kwon (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.07.001) discuss the importance of membrane fusion in immunity-mediating exocytosis and endocytosis, and highlight the contributions of the SNARE proteins in membrane fusion. Although not specifically focused on immunity, the review by Noack and Jaillais (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.06.017) on phosphoinositides provides additional insight into the regulation of membrane trafficking. In the same issue, Isono and Kalinowska (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.07.003 ), Debaux and Vert (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.07.005), and Burkart and Stahl (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.06.016) discuss the role of endosomal sorting complex required for transport (ESCRT) and ubiquitination in endocytosis, as well as the dynamic complexity of plant receptor complexes at the plasma membrane. Even if you don’t find membranes exciting (!), this collection of papers is a must-read for insights into how plant cells perceive and control their environment.
New reviews cover the hot topic of membrane and vesicle trafficking in plant immunity. Gu et al. (Mol. Plant 10.1016/j.molp.2017.07.001) provide an overview of the two membrane trafficking pathways: the secretory pathway involved in movement of antimicrobials, defense proteins and cell wall components outwards, and the endocytic pathway involved in movement of immune receptors inward for signal transduction. They describe the molecular players and regulators, features of these pathways that are unique to plants, how these pathways are involved in defense, and how pathogens sometimes co-opt them to supress immunity. Yun and Kwon (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.07.001) discuss the importance of membrane fusion in immunity-mediating exocytosis and endocytosis, and highlight the contributions of the SNARE proteins in membrane fusion. Although not specifically focused on immunity, the review by Noack and Jaillais (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.06.017) on phosphoinositides provides additional insight into the regulation of membrane trafficking. In the same issue, Isono and Kalinowska (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.07.003 ), Debaux and Vert (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.07.005), and Burkart and Stahl (Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.06.016) discuss the role of endosomal sorting complex required for transport (ESCRT) and ubiquitination in endocytosis, as well as the dynamic complexity of plant receptor complexes at the plasma membrane. Even if you don’t find membranes exciting (!), this collection of papers is a must-read for insights into how plant cells perceive and control their environment.
Opinion: Is there an upper limit to genome size? ($)
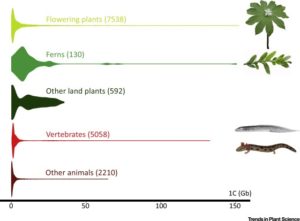 There are only ten organisms known to have genomes larger than 100 Gb in size and six of those are plants. Both Numbers 1 and 2 on the list are plants with genomes that are nearly 50x the size of the human genome (which is 3 Gb), and over 1000x that of Arabidopsis: the 148.8 Gb-genome Paris japonica (Japanese canopy plant), and the 147.3 Gb-genome wisk fern Tmesipteris obliqua. After surveying the size of all known genomes, Hidalgo and Pellicer et al. speculate that 150 Gb could be an upper biological genome limit. Why this would be the limit is less clear, but leading possibilities would seem to be high energy costs involved in DNA replication and repair, and basic cell-size packing constraints. (Summary by Aaron Rashotte). Trends Plant Sci. 10.1016/j.tplants.2017.04.005
There are only ten organisms known to have genomes larger than 100 Gb in size and six of those are plants. Both Numbers 1 and 2 on the list are plants with genomes that are nearly 50x the size of the human genome (which is 3 Gb), and over 1000x that of Arabidopsis: the 148.8 Gb-genome Paris japonica (Japanese canopy plant), and the 147.3 Gb-genome wisk fern Tmesipteris obliqua. After surveying the size of all known genomes, Hidalgo and Pellicer et al. speculate that 150 Gb could be an upper biological genome limit. Why this would be the limit is less clear, but leading possibilities would seem to be high energy costs involved in DNA replication and repair, and basic cell-size packing constraints. (Summary by Aaron Rashotte). Trends Plant Sci. 10.1016/j.tplants.2017.04.005
Letters to the Editor: Does C4 photosynthesis occur in wheat seeds?
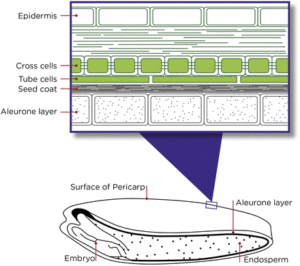 In 2016, Rangan et al. reported on “New evidence for grain specific C4 photosynthesis in wheat,” but later that year Busch and Farquhar responded with “Poor evidence for C4 photosynthesis in the wheat grain.” Now these two groups continue their arguments for and against wheat seed C4 photosynthesis, in a quartet of Letters to the Editor. Is the issue settled? No, but the publication of these letters provides insights into how data are interpreted differently, and how scientific disagreements can be conducted in a open and respectful manner. This is a great topic and collection for students to read and discuss. Plant Physiol. 10.1104/pp.17.00837
In 2016, Rangan et al. reported on “New evidence for grain specific C4 photosynthesis in wheat,” but later that year Busch and Farquhar responded with “Poor evidence for C4 photosynthesis in the wheat grain.” Now these two groups continue their arguments for and against wheat seed C4 photosynthesis, in a quartet of Letters to the Editor. Is the issue settled? No, but the publication of these letters provides insights into how data are interpreted differently, and how scientific disagreements can be conducted in a open and respectful manner. This is a great topic and collection for students to read and discuss. Plant Physiol. 10.1104/pp.17.00837
CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping ($)
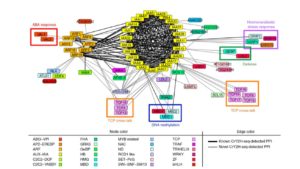 Protein-protein interactions are crucial to our understanding of biology but can be hard to detect. Trigg et al. developed a sophisticated yeast two-hybrid assay augmented with Cre recombinase (CrY2H-seq) to identify the Arabidopsis transcription factor protein-protein interactome. In this method, plasmid libraries carrying transcription factor open-reading frames (ORFs) fused to GAL4 activation domains or DNA-binding domains, flanked by lox sequences (targets of Cre recombinase) are introduced into yeast cells with a genome-integrated Cre recombinase ORF under the control of the GAL7 promoter. Interaction in the yeast cells between the two proteins leads to expression of Cre recombinase, which targets the lox sequences and joins the two plasmids together. Plasmid recovery and sequencing reveals pairs of interacting proteins. Starting with nearly 2000 Arabidopsis transcription factors, the authors identified nearly 9000 novel and known interactions and independently validated hundreds of them. Biological functions implicated by the uncovered interactions include gynoecium development, phosphate sensing and circadian coordination. With CrY2H-seq facilitating massively-multiplexed interactome screening, a more complete arabidopsis interactome as well as potential interactomes for other plants are on the horizon. The searchable dataset ‘Arabidopsis thaliana transcription factor interaction network, version 1’ (AtTFIN-1) is available online (http://signal.salk.edu/interactome/AtTFIN-1.html). Nature Methods 10.1038/nmeth.4343
Protein-protein interactions are crucial to our understanding of biology but can be hard to detect. Trigg et al. developed a sophisticated yeast two-hybrid assay augmented with Cre recombinase (CrY2H-seq) to identify the Arabidopsis transcription factor protein-protein interactome. In this method, plasmid libraries carrying transcription factor open-reading frames (ORFs) fused to GAL4 activation domains or DNA-binding domains, flanked by lox sequences (targets of Cre recombinase) are introduced into yeast cells with a genome-integrated Cre recombinase ORF under the control of the GAL7 promoter. Interaction in the yeast cells between the two proteins leads to expression of Cre recombinase, which targets the lox sequences and joins the two plasmids together. Plasmid recovery and sequencing reveals pairs of interacting proteins. Starting with nearly 2000 Arabidopsis transcription factors, the authors identified nearly 9000 novel and known interactions and independently validated hundreds of them. Biological functions implicated by the uncovered interactions include gynoecium development, phosphate sensing and circadian coordination. With CrY2H-seq facilitating massively-multiplexed interactome screening, a more complete arabidopsis interactome as well as potential interactomes for other plants are on the horizon. The searchable dataset ‘Arabidopsis thaliana transcription factor interaction network, version 1’ (AtTFIN-1) is available online (http://signal.salk.edu/interactome/AtTFIN-1.html). Nature Methods 10.1038/nmeth.4343
Structure of a symmetric photosynthetic reaction center–photosystem ($)
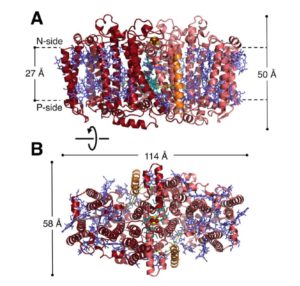 Plants, green algae and cyanobacteria carry out oxygenic photosynthesis through the coordination of two photosystems, PSI and PSII. Many other photosynthetic prokaryotes use a single reaction center to carry out anoxygenic photosynthesis. Gisriel et al. describe the structure of a photosynthetic reaction center from Heliobactrium modesticaldum, “a thermophilic anaerobe isolated from volcanic soil in Iceland.” The novelty of the new structure is that it is composed of a homodimeric reaction center, whereas others are heterodimeric, and is thought to more closely resemble the ancestral state. The authors point out that these bacteria branched early in bacterial evolution, meaning that the reaction center has had a long time to diverge from that of other species, but that its anoxic niche with its similarities to early Earth may have helped it maintain some of the ancestral traits. Science 10.1126/science.aan5611
Plants, green algae and cyanobacteria carry out oxygenic photosynthesis through the coordination of two photosystems, PSI and PSII. Many other photosynthetic prokaryotes use a single reaction center to carry out anoxygenic photosynthesis. Gisriel et al. describe the structure of a photosynthetic reaction center from Heliobactrium modesticaldum, “a thermophilic anaerobe isolated from volcanic soil in Iceland.” The novelty of the new structure is that it is composed of a homodimeric reaction center, whereas others are heterodimeric, and is thought to more closely resemble the ancestral state. The authors point out that these bacteria branched early in bacterial evolution, meaning that the reaction center has had a long time to diverge from that of other species, but that its anoxic niche with its similarities to early Earth may have helped it maintain some of the ancestral traits. Science 10.1126/science.aan5611
Genomic estimation of complex traits reveals ancient maize adaptation to temperate North America
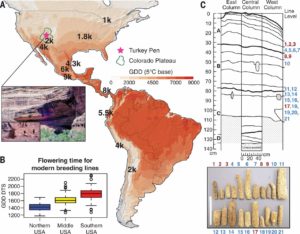 Maize (corn), an important staple of the diet in ancient and modern times, was cultivated at higher altitudes in the southwestern United States, around 2,000 years after its introduction to the lowland US regions. In order to better understand how maize later adapted to high altitudes, authors sequenced the genomes of fifteen 1,900-year-old maize species. Comparison of these sequenced ancient genomes to modern genomes revealed several complex genetic traits, such as an earlier flowering time, that allowed ancient maize to adapt to shorter growing seasons at higher altitudes. Understanding the genetic traits that helped ancient maize to adapt to environmental changes can inform plant breeders as to how to adapt maize to the changing global climate. (Summary by Tyra McCray) Science 10.1126/science.aam9425.
Maize (corn), an important staple of the diet in ancient and modern times, was cultivated at higher altitudes in the southwestern United States, around 2,000 years after its introduction to the lowland US regions. In order to better understand how maize later adapted to high altitudes, authors sequenced the genomes of fifteen 1,900-year-old maize species. Comparison of these sequenced ancient genomes to modern genomes revealed several complex genetic traits, such as an earlier flowering time, that allowed ancient maize to adapt to shorter growing seasons at higher altitudes. Understanding the genetic traits that helped ancient maize to adapt to environmental changes can inform plant breeders as to how to adapt maize to the changing global climate. (Summary by Tyra McCray) Science 10.1126/science.aam9425.
In vivo FRET–FLIM reveals cell-type-specific protein interactions in Arabidopsis roots ($)
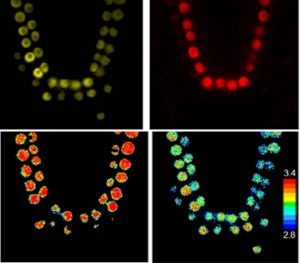 Long et al. examined transcription factor complex formation in vivo in Arabidopsis roots using a technique that combines FRET (Förster Resonance Energy Transfer) and FLIM (Fluorescence Lifetime imaging Microscopy). Using this method, the authors were able to observe cell-type specific complex formation between transcription factors known to specify root development, specifically, SHORT-ROOT (SHR), SCARECROW (SCR). JACKDAW (JKD), and NUTCRACKER (NUC). The authors found that protein complexes can change their conformation in a cell-type dependent manner to regulate gene expression leading to precise specification and maintenance of particular cell fates within the Arabidopsis root meristem. Nature 10.1038/nature23317
Long et al. examined transcription factor complex formation in vivo in Arabidopsis roots using a technique that combines FRET (Förster Resonance Energy Transfer) and FLIM (Fluorescence Lifetime imaging Microscopy). Using this method, the authors were able to observe cell-type specific complex formation between transcription factors known to specify root development, specifically, SHORT-ROOT (SHR), SCARECROW (SCR). JACKDAW (JKD), and NUTCRACKER (NUC). The authors found that protein complexes can change their conformation in a cell-type dependent manner to regulate gene expression leading to precise specification and maintenance of particular cell fates within the Arabidopsis root meristem. Nature 10.1038/nature23317
Metabolic engineering of anthocyanin and betalain pigments for health and aesthetics: Purple rice, blue chrysanthemums and violet tomatoes
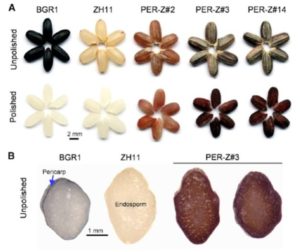 Pigment engineering was featured in three recent papers. Anthocyanins are blue pigments valued for their antioxidant health benefits and for their beauty, but their biosynthesis and chemistry is complex. Noda et al. introduced two genes to produce blue anthocyanins in chrysanthemum petals (Sci Advances 10.1126/sciadv,1602785). They found that the blue pigment formed as a copigment arising from the molecular interaction between 3′,5′-diglucosylated delphinidin and flavone glucosides. Zhu et al. describe efforts to engineer anthocyanin biosynthesis into rice endosperm. They achieved this goal by introducing eight genes (two regulatory and six structural) under the control of endosperm-specific promoters. In transgenic plants, the authors also observed enhanced expression of six endogenous anthocyanin biosynthetic genes that are normally silent in the endosperm. (Mol. Plant 10.1016/j.molp.2017.05.008). Polturak et al. describe engineering betalain (red betacyanins and yellow betaxanthins) production in several plants. Like anthocyanins, betalains have antioxidant properties. Unexpectedly, the authors found that enhanced betalain production in tobacco led to resistance to Botrytis cinerea (gray mold). (Proc. Natl. Acad. Sci. USA 10.1073/pnas.1707176114).
Pigment engineering was featured in three recent papers. Anthocyanins are blue pigments valued for their antioxidant health benefits and for their beauty, but their biosynthesis and chemistry is complex. Noda et al. introduced two genes to produce blue anthocyanins in chrysanthemum petals (Sci Advances 10.1126/sciadv,1602785). They found that the blue pigment formed as a copigment arising from the molecular interaction between 3′,5′-diglucosylated delphinidin and flavone glucosides. Zhu et al. describe efforts to engineer anthocyanin biosynthesis into rice endosperm. They achieved this goal by introducing eight genes (two regulatory and six structural) under the control of endosperm-specific promoters. In transgenic plants, the authors also observed enhanced expression of six endogenous anthocyanin biosynthetic genes that are normally silent in the endosperm. (Mol. Plant 10.1016/j.molp.2017.05.008). Polturak et al. describe engineering betalain (red betacyanins and yellow betaxanthins) production in several plants. Like anthocyanins, betalains have antioxidant properties. Unexpectedly, the authors found that enhanced betalain production in tobacco led to resistance to Botrytis cinerea (gray mold). (Proc. Natl. Acad. Sci. USA 10.1073/pnas.1707176114).
Systemic transport of trans-zeatin and its precursor have differing roles in Arabidopsis shoots
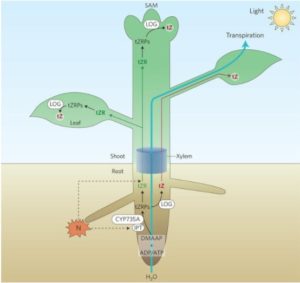 Plant hormones are made in one tissue and usually transported to act in another. One example of this is the transport of cytokinin. In the root, the precursor trans-zeatin riboside (tZR) is synthesized, then xylem loaded and transported to the shoot. Once at a site of action like the leaf or shoot apical meristem (SAM), inactive tZR is converted by the LONELY GUY enzyme into active trans-zeatin (tZ) to function in cytokinin-related processes. Osugi et al. used radio-labeled cytokinins and grafting experiments with multiple Arabidopsis mutants including cytokinin transport mutants log1,2,3,4,5,7,8 and biosynthesis mutants ipt3,5,7 to show that the active tZ form is also transported and functionally competent in cytokinin response in leaves, but not the SAM. This occurs independently of the previously demonstrated tZR transport to both leaves and SAM. While it is unclear why both modes occur, it suggest a new and more complicated model for the transport and function of cytokinin forms to regulate leaf and SAM growth. (Summary by Aaron Rashotte) Nature Plants 10.1038/nplants.2017.112
Plant hormones are made in one tissue and usually transported to act in another. One example of this is the transport of cytokinin. In the root, the precursor trans-zeatin riboside (tZR) is synthesized, then xylem loaded and transported to the shoot. Once at a site of action like the leaf or shoot apical meristem (SAM), inactive tZR is converted by the LONELY GUY enzyme into active trans-zeatin (tZ) to function in cytokinin-related processes. Osugi et al. used radio-labeled cytokinins and grafting experiments with multiple Arabidopsis mutants including cytokinin transport mutants log1,2,3,4,5,7,8 and biosynthesis mutants ipt3,5,7 to show that the active tZ form is also transported and functionally competent in cytokinin response in leaves, but not the SAM. This occurs independently of the previously demonstrated tZR transport to both leaves and SAM. While it is unclear why both modes occur, it suggest a new and more complicated model for the transport and function of cytokinin forms to regulate leaf and SAM growth. (Summary by Aaron Rashotte) Nature Plants 10.1038/nplants.2017.112
Rhamnose-containing cell wall polymers suppress helical plant growth independently of microtubule orientation
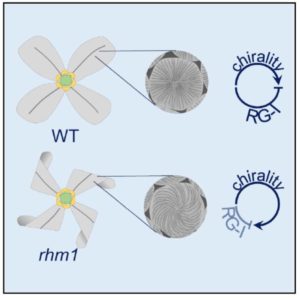 Saffer et al. identified an Arabidpsis mutant with swirled petals and with petal epidermal cells that show a left-handed (but never right-handed) twist. They mapped the mutation to the RHAMNOSE BIOSYNTHESIS1 (RHM1) gene, which is most highly expressed in petal epidermal cells and encodes an enzyme that catalyzes the biosynthesis of UDP-L-rhamnose, a component of pectin, indicating that pectin serves to suppress twisting. The helical twisting does not involve the cytoskeleton, and appears to derive from an inherently chiral component of the cell wall. Curr. Biol. 10.1016/j.cub.2017.06.032
Saffer et al. identified an Arabidpsis mutant with swirled petals and with petal epidermal cells that show a left-handed (but never right-handed) twist. They mapped the mutation to the RHAMNOSE BIOSYNTHESIS1 (RHM1) gene, which is most highly expressed in petal epidermal cells and encodes an enzyme that catalyzes the biosynthesis of UDP-L-rhamnose, a component of pectin, indicating that pectin serves to suppress twisting. The helical twisting does not involve the cytoskeleton, and appears to derive from an inherently chiral component of the cell wall. Curr. Biol. 10.1016/j.cub.2017.06.032
LAZY1 family contributes to gravity signaling within statocytes and branch angle control of roots and shoots
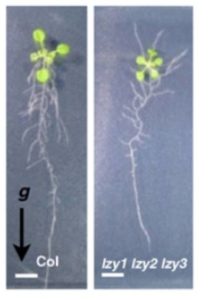 It’s easy to demonstrate that plants sense gravity, and we also know that statocytes are involved in the perception of gravity. Statocytes are gravity-sensing cells that contain dense starch-containing amyloplasts that move within the cell in the direction of gravity. Differential growth to accommodate a change in gravity orientation involves auxin transport, but the steps between statocyte movement and auxin transport have been difficult to define. Taniguchi et al. showed that in LAZY triple mutants (lzy1 lzy2 lzy3), statocyote redistribution occurs normally but the redistribution of auxin transporter (PIN) proteins is affected, which demonstrates that the LZY family transduces information between statocytes and auxin transport. Plant Cell 10.1105/tpc.16.00575
It’s easy to demonstrate that plants sense gravity, and we also know that statocytes are involved in the perception of gravity. Statocytes are gravity-sensing cells that contain dense starch-containing amyloplasts that move within the cell in the direction of gravity. Differential growth to accommodate a change in gravity orientation involves auxin transport, but the steps between statocyte movement and auxin transport have been difficult to define. Taniguchi et al. showed that in LAZY triple mutants (lzy1 lzy2 lzy3), statocyote redistribution occurs normally but the redistribution of auxin transporter (PIN) proteins is affected, which demonstrates that the LZY family transduces information between statocytes and auxin transport. Plant Cell 10.1105/tpc.16.00575
Stem parasitic plant Cuscuta australis (dodder) transfers herbivory-induced signals among plants
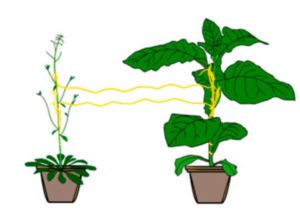 Parasitic plants such as Cuscuta astralis (dodder) form connections with their host plants through which nutrients and other molecules pass. Using mutant plants and transcriptomic assays, Hettenhausen and Li et al. showed that two or more plants connected by Cuscuta bridges shared information through these bridges. Specifically, when one host plant was wounded or subjected to insect herbivory, another plant connected through the Cuscuta bridge showed rapid transcriptional responses. Information is likely to include both jasmonate and other components. This study shows that plant-to-plant information can be transmitted through parasitic plant networks. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1704536114
Parasitic plants such as Cuscuta astralis (dodder) form connections with their host plants through which nutrients and other molecules pass. Using mutant plants and transcriptomic assays, Hettenhausen and Li et al. showed that two or more plants connected by Cuscuta bridges shared information through these bridges. Specifically, when one host plant was wounded or subjected to insect herbivory, another plant connected through the Cuscuta bridge showed rapid transcriptional responses. Information is likely to include both jasmonate and other components. This study shows that plant-to-plant information can be transmitted through parasitic plant networks. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1704536114




Leave a Reply
Want to join the discussion?Feel free to contribute!