What We’re Reading: August 10
Genome assemblies of maize lines Mo17 and W22: Extensive intraspecific variation, and resource for functional biology
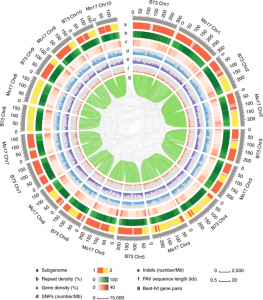 The maize genome is largely composed of transposable elements, which is one reason maize has been such a powerful genetic model. However, these transposons also mean that there is a great deal of genetic variability between inbred lines, which can contribute to heterosis (hybrid vigor). In a pair of papers, Sun et al. report a de novo genome assembly of Mo17, which is widely used as a male parent in hybrid production, and Springer et al. report the assembly of a reference genome for W22, which is widely used for transposon-based gene mapping. These new genome assemblies, which join those of maize lines B73 and PH207, provide unprecedented insights into the variations of gene order, structure, and transposon landscape between maize inbreds. (Summary by Mary Williams) Nature Genetics 10.1038/s41588-018-0182-0 and 10.1038/s41588-018-0158-0
The maize genome is largely composed of transposable elements, which is one reason maize has been such a powerful genetic model. However, these transposons also mean that there is a great deal of genetic variability between inbred lines, which can contribute to heterosis (hybrid vigor). In a pair of papers, Sun et al. report a de novo genome assembly of Mo17, which is widely used as a male parent in hybrid production, and Springer et al. report the assembly of a reference genome for W22, which is widely used for transposon-based gene mapping. These new genome assemblies, which join those of maize lines B73 and PH207, provide unprecedented insights into the variations of gene order, structure, and transposon landscape between maize inbreds. (Summary by Mary Williams) Nature Genetics 10.1038/s41588-018-0182-0 and 10.1038/s41588-018-0158-0
Footprints of parasitism in the genome of the parasitic flowering plant Cuscuta campestris
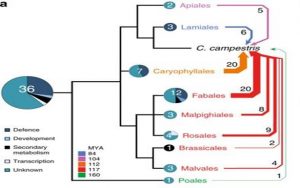 Even without knowing a lot about parasitic plants, you can probably guess some of the insights that come from the first parasitic plant genomic sequence. Because parasitic plants get their nutrients from another organism (functionally, they become heterotrophic), you might expect they would gradually lose the genes needed for nutrient uptake and for photosynthesis. You would be right; Vogel et al. show exactly that with their analysis of the genome of Cuscuta campestris (dodder). They also give evidence that dodder has acquired some genes horizontally, through its intimate association with its hosts. The dodder genome includes “64 novel high confidence horizontal gene transfer candidates from at least 32 different donor sequences,” and many of these genes were shown to be transcribed and upregulated at the site where the parasite interfaces with the host, suggesting a role in the infection process. (Summary by Mary Williams) Nature Comms. 10.1038/s41467-018-04344-z
Even without knowing a lot about parasitic plants, you can probably guess some of the insights that come from the first parasitic plant genomic sequence. Because parasitic plants get their nutrients from another organism (functionally, they become heterotrophic), you might expect they would gradually lose the genes needed for nutrient uptake and for photosynthesis. You would be right; Vogel et al. show exactly that with their analysis of the genome of Cuscuta campestris (dodder). They also give evidence that dodder has acquired some genes horizontally, through its intimate association with its hosts. The dodder genome includes “64 novel high confidence horizontal gene transfer candidates from at least 32 different donor sequences,” and many of these genes were shown to be transcribed and upregulated at the site where the parasite interfaces with the host, suggesting a role in the infection process. (Summary by Mary Williams) Nature Comms. 10.1038/s41467-018-04344-z
Genome-scale sequence disruption following biolistic transformation in rice and maize
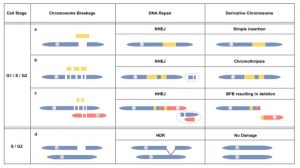 The two classic approaches to introducing DNA into a plant’s genome are by harnessing Agrobacterium tumefaciens’ fascinating gene-transfer skill, or by shooting the new DNA into the cell using a “biolistic” (gun) approach. Because of Agrobacterium’s restricted choice of hosts, the biolistic method has been preferred for monocots. Traditionally, regenerants are characterized phenotypically and by DNA hybridization, so the broader genomic effects of the biolistic bombardment aren’t well known. To find out, Liu et al. bombarded linear (phage lambda) and circular DNA into rice and wheat, and then analyzed the resulting lines using whole-genome sequencing. They found “multiple forms of genome damage …. including chromosome breakage and shattering and extreme copy number variation.” The authors speculate that the outcome may depend on the stage of the cell cycle when the event occurred. The authors also observe that “there is no evidence that a genomic rearrangement has compromised the safety of a plant used as food” and “there is a rigorous selection process that eliminates deleterious mutations during the breeding process.” (Summary by Mary Williams) bioRxiv 10.1101/379644
The two classic approaches to introducing DNA into a plant’s genome are by harnessing Agrobacterium tumefaciens’ fascinating gene-transfer skill, or by shooting the new DNA into the cell using a “biolistic” (gun) approach. Because of Agrobacterium’s restricted choice of hosts, the biolistic method has been preferred for monocots. Traditionally, regenerants are characterized phenotypically and by DNA hybridization, so the broader genomic effects of the biolistic bombardment aren’t well known. To find out, Liu et al. bombarded linear (phage lambda) and circular DNA into rice and wheat, and then analyzed the resulting lines using whole-genome sequencing. They found “multiple forms of genome damage …. including chromosome breakage and shattering and extreme copy number variation.” The authors speculate that the outcome may depend on the stage of the cell cycle when the event occurred. The authors also observe that “there is no evidence that a genomic rearrangement has compromised the safety of a plant used as food” and “there is a rigorous selection process that eliminates deleterious mutations during the breeding process.” (Summary by Mary Williams) bioRxiv 10.1101/379644
A single cis-element that controls cell-type specific expression in Arabidopsis
 Multicellular organisms have different tissues that carry out diverse and specialized functions, and tissue-specific expression is the feature that gives each tissue its specific protein content. Despite its importance, the mechanisms that control spatial patterning is poorly understood. In this work, Kneřová et al. focus their investigation in the transcription factor MYB76 that is expressed specifically in bundle sheath cells. Kneřová et al. found that a seven-nucleotide portion of the MYB76 promoter is necessary and sufficient to direct tissue specific gene expression, and that oligomerization of this portion increases the expression level of the regulated transcript. The authors propose the use of this very short promoter in synthetic biology, as short promoters have the advantage of reduced silencing and leakiness or off-target expression. Driving expression of various genes speicifically in the bundle sheath cells could facilitate the control of: storage carbohydrates and ions, hydraulic conductance, transport of metabolites in and out of veins, responses to high light episodes, and lead to photosynthetic enhancements. (Summary by Aime Jaskolowski) bioRxiv
Multicellular organisms have different tissues that carry out diverse and specialized functions, and tissue-specific expression is the feature that gives each tissue its specific protein content. Despite its importance, the mechanisms that control spatial patterning is poorly understood. In this work, Kneřová et al. focus their investigation in the transcription factor MYB76 that is expressed specifically in bundle sheath cells. Kneřová et al. found that a seven-nucleotide portion of the MYB76 promoter is necessary and sufficient to direct tissue specific gene expression, and that oligomerization of this portion increases the expression level of the regulated transcript. The authors propose the use of this very short promoter in synthetic biology, as short promoters have the advantage of reduced silencing and leakiness or off-target expression. Driving expression of various genes speicifically in the bundle sheath cells could facilitate the control of: storage carbohydrates and ions, hydraulic conductance, transport of metabolites in and out of veins, responses to high light episodes, and lead to photosynthetic enhancements. (Summary by Aime Jaskolowski) bioRxiv
Divergent strategies to maintain steady state protein levels in algal and angiosperm chloroplasts
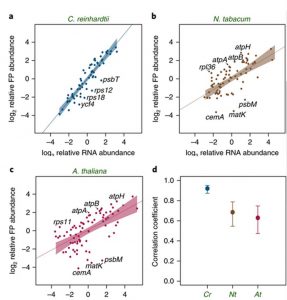 Plants and their algal predecessors both utilize chloroplasts for photosynthesis and other essential metabolic processes. The endosymbiotic origin and subsequent co-evolution of this autotrophic organelle introduced striking regulatory mechanisms combining both prokaryotic and eukaryotic strategies for optimal functionality and control. In a recent article, Trosch et al. (Nature Plants) performed a comprehensive comparison of chloroplast transcription and translation in a model chlorophyte alga (Chlamydomonas reinhardtii) and two model angiosperms (Arabidopsis thaliana and Nicotiana benthamiana) to assess the conservation of regulatory mechanisms in chloroplasts. Through a combination of chloroplast ribosome profiling and transcriptomics, the authors discovered a high similarity in steady state protein levels despite significant differences in transcriptional abundances across these organisms. Together, their work argues that these divergent lineages have developed distinct strategies to maintain protein synthesis in chloroplasts. (Summary by Phil Carella) Nature Plants 10.1038/s41477-018-0211-0
Plants and their algal predecessors both utilize chloroplasts for photosynthesis and other essential metabolic processes. The endosymbiotic origin and subsequent co-evolution of this autotrophic organelle introduced striking regulatory mechanisms combining both prokaryotic and eukaryotic strategies for optimal functionality and control. In a recent article, Trosch et al. (Nature Plants) performed a comprehensive comparison of chloroplast transcription and translation in a model chlorophyte alga (Chlamydomonas reinhardtii) and two model angiosperms (Arabidopsis thaliana and Nicotiana benthamiana) to assess the conservation of regulatory mechanisms in chloroplasts. Through a combination of chloroplast ribosome profiling and transcriptomics, the authors discovered a high similarity in steady state protein levels despite significant differences in transcriptional abundances across these organisms. Together, their work argues that these divergent lineages have developed distinct strategies to maintain protein synthesis in chloroplasts. (Summary by Phil Carella) Nature Plants 10.1038/s41477-018-0211-0
Predominant Golgi-residency of the plant K/HDEL receptor is essential for its function in mediating ER retention
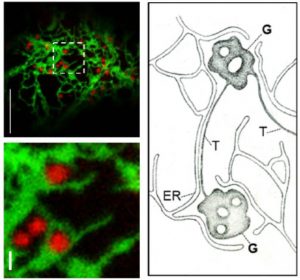 Cells use a sophisticated sorting system to ensure that proteins get to the proper destination. Proteins that are supposed to stay in the ER carry special tags (KDEL or HDEL, recognized by the ERD2 receptor) that prevent them from being swept along with other proteins out of the ER and into the Golgi. Silva-Alvim et al. examined ERD2 function in plants. A key finding is that the position of the fluorescent tag determines ERD2 function, and previous models have been built on studies in which the presence of the tag causes the protein to be inactive. One model for ERD2 function is that it catches K/HDEL proteins that have escaped into the Golgi and carries them in a retrograde direction back to the ER. Interestingly, the authors were unable to detect any ERD2 in the ER, suggesting that it is not directly involved in retrograde transport, or that it leaves the ER very rapidly. As the authors observe, “one of the most conserved steps in the secretory pathway is one of the least understood,” but the tools and assays developed in this work will help to sort things out. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00426
Cells use a sophisticated sorting system to ensure that proteins get to the proper destination. Proteins that are supposed to stay in the ER carry special tags (KDEL or HDEL, recognized by the ERD2 receptor) that prevent them from being swept along with other proteins out of the ER and into the Golgi. Silva-Alvim et al. examined ERD2 function in plants. A key finding is that the position of the fluorescent tag determines ERD2 function, and previous models have been built on studies in which the presence of the tag causes the protein to be inactive. One model for ERD2 function is that it catches K/HDEL proteins that have escaped into the Golgi and carries them in a retrograde direction back to the ER. Interestingly, the authors were unable to detect any ERD2 in the ER, suggesting that it is not directly involved in retrograde transport, or that it leaves the ER very rapidly. As the authors observe, “one of the most conserved steps in the secretory pathway is one of the least understood,” but the tools and assays developed in this work will help to sort things out. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00426
Chloroplast biogenesis controlled by DELLA-TOC159 interaction in early plant development
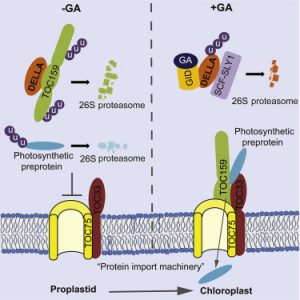 Plant photosynthesis occurs in the chloroplast, the green organelles that are the most famous members of the plastid family. Chloroplast biogenesis starts with illumination at germination, when the colorles, non-photosynthetic proplastid acquires photosynthetic activity as it greens. Gaining photosynthetic function depends on the import of thousands of different preproteins into the developing organelle through the chloroplast protein import machinery. This machinery comprises TOC75, the protein-conducting channel, and the import receptors TOC33 and TOC159. It has been previously demonstrated that gibberellic acid (GA)-dependent downregulation of DELLA proteins is essential for chlorophyll and carotenoid biosynthesis. In this work Shanmugabalaji et al. demonstrate that the expression of the DELLA protein RGL2 is inversely correlated with expression of TOC159 during seed germination. They show that RGL2 and TOC159 physically interact in yeast an in planta and that the inhibition of the ubiquitin/proteasome system increases TOC159 under low-GA conditions. Finally, they conclude that direct interaction with DELLA promotes the degradation of the TOC159 import receptor; upon destruction of DELLA, TOC159 is immediately available for assembly into the TOC complex, which allows full import of photosynthesis-associated proteins into the chloroplast. (Summary by Aime Jaskolowski) Curr. Biol. 10.1016/j.cub.2018.06.006
Plant photosynthesis occurs in the chloroplast, the green organelles that are the most famous members of the plastid family. Chloroplast biogenesis starts with illumination at germination, when the colorles, non-photosynthetic proplastid acquires photosynthetic activity as it greens. Gaining photosynthetic function depends on the import of thousands of different preproteins into the developing organelle through the chloroplast protein import machinery. This machinery comprises TOC75, the protein-conducting channel, and the import receptors TOC33 and TOC159. It has been previously demonstrated that gibberellic acid (GA)-dependent downregulation of DELLA proteins is essential for chlorophyll and carotenoid biosynthesis. In this work Shanmugabalaji et al. demonstrate that the expression of the DELLA protein RGL2 is inversely correlated with expression of TOC159 during seed germination. They show that RGL2 and TOC159 physically interact in yeast an in planta and that the inhibition of the ubiquitin/proteasome system increases TOC159 under low-GA conditions. Finally, they conclude that direct interaction with DELLA promotes the degradation of the TOC159 import receptor; upon destruction of DELLA, TOC159 is immediately available for assembly into the TOC complex, which allows full import of photosynthesis-associated proteins into the chloroplast. (Summary by Aime Jaskolowski) Curr. Biol. 10.1016/j.cub.2018.06.006
Minimum requirements for changing and maintaining endodermis cell identity in the Arabidopsis root ($)
 Forward-genetic, loss-of-function studies have been invaluable in identifying factors necessary for the formation of the root endodermis. Drapek et al. used an ectopic expression approach to identify factors that are sufficient to produce endodermal-like cells in different root regions. They found that the combination of the key transcription factor SHR and the peptide ligand CIF2 is sufficient (in some conditions) to produce endodermis-like transcriptional and cellular responses in the subepidermal cell layers. The authors conclude that CIF2 is needed to stabilize endodermal identity. Furthermore, they showed that both positional and lineage information is required for cell fate specification.(Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0213-y
Forward-genetic, loss-of-function studies have been invaluable in identifying factors necessary for the formation of the root endodermis. Drapek et al. used an ectopic expression approach to identify factors that are sufficient to produce endodermal-like cells in different root regions. They found that the combination of the key transcription factor SHR and the peptide ligand CIF2 is sufficient (in some conditions) to produce endodermis-like transcriptional and cellular responses in the subepidermal cell layers. The authors conclude that CIF2 is needed to stabilize endodermal identity. Furthermore, they showed that both positional and lineage information is required for cell fate specification.(Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0213-y
HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers ($)
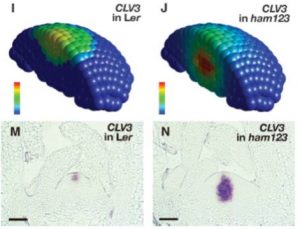 The interaction between transcription factors WUSCHEL (WUS) and CLAVATA3 (CLV3) controls the size of the meristem. Although WUS is known to activate CLV3 expression, their expression domains don’t fully overlap, with CLV3 being restricted to the upper part of the meristem. Previously, in the multiple ham mutant (loss of function of HAIRY MERISTEM family) CLV3 expression was observed in the lower, rib meristem. Using computational and experimental approaches, Zhou et al. showed that WUS activates CLV3 only in the absence of HAM. Their model faithfully predicts the expression domain of CLV3 in a variety of mutant backgrounds, and during de novo meristem formation in leaf axils. (Summary by Mary Williams) Science 10.1126/science.aar8638
The interaction between transcription factors WUSCHEL (WUS) and CLAVATA3 (CLV3) controls the size of the meristem. Although WUS is known to activate CLV3 expression, their expression domains don’t fully overlap, with CLV3 being restricted to the upper part of the meristem. Previously, in the multiple ham mutant (loss of function of HAIRY MERISTEM family) CLV3 expression was observed in the lower, rib meristem. Using computational and experimental approaches, Zhou et al. showed that WUS activates CLV3 only in the absence of HAM. Their model faithfully predicts the expression domain of CLV3 in a variety of mutant backgrounds, and during de novo meristem formation in leaf axils. (Summary by Mary Williams) Science 10.1126/science.aar8638
Natural variation in OsLG3 increases drought tolerance in rice
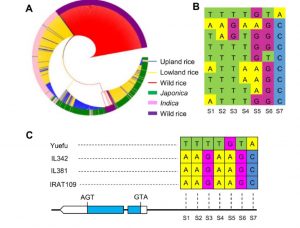 Through a GWAS study, Yu, Xiong et al. previously identified a transcription-factor encoding gene, OsLG3, with alleles that contribute to increased grain length in rice. In this new work, Xiong, Yu et al. showed that the same alleles of OsLG3 that confers longer grain length also enhance drought tolerance. By surveying a panel of varieties, the authors were able to show that several SNPs correlate with enhanced germination in osmotic stress, and that these SNPs are located in the promoter and positively affect OsLG3 transcription. Further studies confirmed that elevated OsLG3 expression correlates with drought tolerance, possibly through “transcriptional regulation of stress-related and ROS-scavenging–related genes.” (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.01492
Through a GWAS study, Yu, Xiong et al. previously identified a transcription-factor encoding gene, OsLG3, with alleles that contribute to increased grain length in rice. In this new work, Xiong, Yu et al. showed that the same alleles of OsLG3 that confers longer grain length also enhance drought tolerance. By surveying a panel of varieties, the authors were able to show that several SNPs correlate with enhanced germination in osmotic stress, and that these SNPs are located in the promoter and positively affect OsLG3 transcription. Further studies confirmed that elevated OsLG3 expression correlates with drought tolerance, possibly through “transcriptional regulation of stress-related and ROS-scavenging–related genes.” (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.01492
OST1 activation by the brassinosteroid-regulated kinase CDG1-Like 1 in stomatal closure ($)
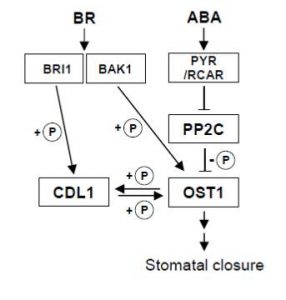 As gatekeepers between the outside world and the interior of the leaf, guard cells are extraordinarily sensitive to their environment. Closing keeps out pathogens and keeps in water vapor, but opening is needed to admit CO2. The web of interactions that control stomatal aperture are also correspondingly complex. Although ABA is well known as a regulator of stomatal aperture, the role of BRs is less well understood. Kim et al. have identified a new convergence point between ABA and brassinosteroids in stomatal aperture. They show that the BR-regulated kinase CDG1-LIKE 1 (CDL1) interacts with and phosphorylates OST1, a key regulator of guard cell closure. They also show that both ABA and BR induce transphosphorylation between OST1 and CDL1. How this interaction fits in with BRs’ growth-regulatory functions remains to be determined. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00239
As gatekeepers between the outside world and the interior of the leaf, guard cells are extraordinarily sensitive to their environment. Closing keeps out pathogens and keeps in water vapor, but opening is needed to admit CO2. The web of interactions that control stomatal aperture are also correspondingly complex. Although ABA is well known as a regulator of stomatal aperture, the role of BRs is less well understood. Kim et al. have identified a new convergence point between ABA and brassinosteroids in stomatal aperture. They show that the BR-regulated kinase CDG1-LIKE 1 (CDL1) interacts with and phosphorylates OST1, a key regulator of guard cell closure. They also show that both ABA and BR induce transphosphorylation between OST1 and CDL1. How this interaction fits in with BRs’ growth-regulatory functions remains to be determined. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00239
The fate of scent in insect- vs. wind-pollinated flowers ($)
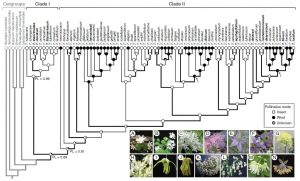 Flower traits (mainly flower colour, position, shape, size, reward and scent) in present-day plants are the result of past selection pressures. Among these traits, scent is a crucial component mediating pollinator attraction, and is often greatly reduced in abiotic-pollinated plants. Wang et al. characterize the volatile organic compounds (VOCs) in very closely-related species from the genus Thalictrum that are either insect- or wind- pollinated. Their results show that there is no clear scent ‘signature’ of a pollination mode, and the VOC profiles were independent of phylogenetic relationships. However, the VOCs of insect-pollinated species were able to elicit a response in potential pollinators while VOCs of wind-pollinated species failed to do so. In conclusion, the authors propose a scenario where the ancestral pollination mode of the genus is insect-mediated and some species transitioned to wind pollination. In insect-pollinated species, the scent’s capacity to attract pollinators is maintained by pollinator-mediated selection. When the studied species transitioned to a wind pollination, this selective pressure was released, and scent lost its ability to induce pollinator responses. (Summary by Mariana Antonietti) Ann. Bot. 10.1093/aob/mcy131
Flower traits (mainly flower colour, position, shape, size, reward and scent) in present-day plants are the result of past selection pressures. Among these traits, scent is a crucial component mediating pollinator attraction, and is often greatly reduced in abiotic-pollinated plants. Wang et al. characterize the volatile organic compounds (VOCs) in very closely-related species from the genus Thalictrum that are either insect- or wind- pollinated. Their results show that there is no clear scent ‘signature’ of a pollination mode, and the VOC profiles were independent of phylogenetic relationships. However, the VOCs of insect-pollinated species were able to elicit a response in potential pollinators while VOCs of wind-pollinated species failed to do so. In conclusion, the authors propose a scenario where the ancestral pollination mode of the genus is insect-mediated and some species transitioned to wind pollination. In insect-pollinated species, the scent’s capacity to attract pollinators is maintained by pollinator-mediated selection. When the studied species transitioned to a wind pollination, this selective pressure was released, and scent lost its ability to induce pollinator responses. (Summary by Mariana Antonietti) Ann. Bot. 10.1093/aob/mcy131



