Updating plant ribosomal protein nomenclature
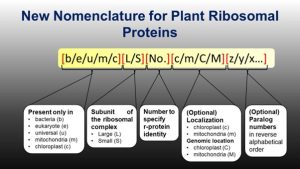 The ribosome is a macromolecular structure made up of many subunits. The proteins in these subunits were named based on their sedimentation rates and gel mobility, but different groups gave the proteins different names in various species, not based on homology. A ribosomal protein (r-protein) nomenclature system was proposed that indicates the origin of the r-protein (e.g., b for bacteria, e for eukaryote, etc.), but it is not adequate for plants as, unlike animals, plant organelle genomes also encode r-proteins, and plants carry multiple genes encoding the same r-proteins. Now Scarpin et al. propose an upgrade to the system of plant r-protein nomenclature. Specifically, they propose to (1) add a suffix, ‘c’ or ‘m’, for chloroplast and mitochondrial-targeted r-proteins, (2) capitalize ‘C’ or ‘M’ if the gene encoding that protein is encoded in respective organelle genome and (3) add a suffix starting from the end of alphabet (z, y, x and so on) for paralogs. These upgrades are proposed based on feedback received on a preprint, social media, email communications, and discussions at the Plant Biology 2022 conference. (Summary by Kamal Kumar Malukani, @KamalMalukani) Plant Cell 10.1093/plcell/koac333
The ribosome is a macromolecular structure made up of many subunits. The proteins in these subunits were named based on their sedimentation rates and gel mobility, but different groups gave the proteins different names in various species, not based on homology. A ribosomal protein (r-protein) nomenclature system was proposed that indicates the origin of the r-protein (e.g., b for bacteria, e for eukaryote, etc.), but it is not adequate for plants as, unlike animals, plant organelle genomes also encode r-proteins, and plants carry multiple genes encoding the same r-proteins. Now Scarpin et al. propose an upgrade to the system of plant r-protein nomenclature. Specifically, they propose to (1) add a suffix, ‘c’ or ‘m’, for chloroplast and mitochondrial-targeted r-proteins, (2) capitalize ‘C’ or ‘M’ if the gene encoding that protein is encoded in respective organelle genome and (3) add a suffix starting from the end of alphabet (z, y, x and so on) for paralogs. These upgrades are proposed based on feedback received on a preprint, social media, email communications, and discussions at the Plant Biology 2022 conference. (Summary by Kamal Kumar Malukani, @KamalMalukani) Plant Cell 10.1093/plcell/koac333