Update: Cytoplasmic mRNA dynamics
 By
By
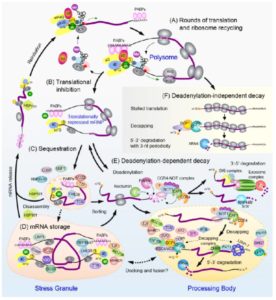
The export of an mRNA from the nucleus to the cytoplasm begins an odyssey of dynamic regulation that determines the location, longevity and use of the transcript in the production of polypeptides by ribosomes in plant cells. Recent leveraging of mutants, imaging of fluorescent proteins and ‘omics’ of protein and ribosome association with mRNAs have significantly enriched our understanding of the intricate regulation of the fates of transcripts within the cytoplasm of plant cells. This Update explores the connections between translation, decay and storage of mRNAs that involve three heterogeneous mRNA-ribonucleoprotein (mRNP) complexes: polyribosomes (polysomes), processing bodies (PBs) and stress granules (SGs) (Box 1). We begin with a brief overview of cellular surveillance of mRNA integrity during the first (pioneer) round of translation. Next, we consider mRNA decay initiated on translating ribosomes, including co-translational exonucleolytic decay and micro(mi)RNA-targeted decay. Then we detail the general decay process of mRNAs initiated by enzymatic deadenylation of the 3’ poly(A) tail and removal of the 5’-742 mG-cap. Finally, we discuss the selective sequestration of translationally repressed mRNAs in SGs under severe environmental stress. This dynamic interplay between mRNA translation, stabilization and turnover fine-tunes the differential regulation of genes to enable developmental plasticity and effective environmental responses.