Translation has something to say in nitrogen-fixing symbiosis
Traubenik et al. highlight the importance of translational control and mRNA decay for successful establishment of the nitrogen-fixing symbiosis. Plant Cell https://doi.org/10.1105/tpc.19.00647
By Soledad Traubenik, Mauricio Reynoso, Flavio Blanco, and María Eugenia Zanetti
Background: Legume plants have the capacity to establish a nitrogen-fixing symbiosis with soil bacteria, which has important implications in nature and agriculture. Plant cells engaged in this symbiosis are reprogrammed to form a new lateral organ in the root called a nodule. This process occurs via tightly regulated mechanisms that alter gene expression at multiple levels. Transcription of certain genomic regions produces new RNA molecules. Some RNAs are translated into proteins, the so-called coding RNAs, while others act as regulatory non-coding RNA molecules that control transcription, processing, translation and/or elimination of RNA molecules. Previous studies illuminated how RNA abundance is modified during nitrogen-fixing symbiosis; however, we know very little about the mechanisms that affects processing, translatability and stability of RNA molecules during the symbiosis process.
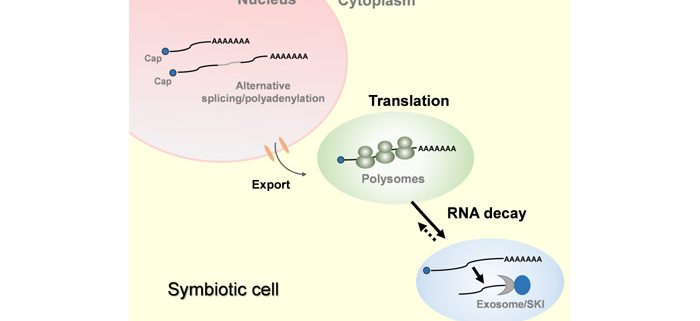
 Question: We investigated how RNA translation affects global changes in gene expression during symbiosis. We also asked whether these regulatory mechanisms operate in all cells committed for symbiosis or in specific cells. Finally, we addressed whether differences in RNA processing might influence RNA translatability and/or stability.
Question: We investigated how RNA translation affects global changes in gene expression during symbiosis. We also asked whether these regulatory mechanisms operate in all cells committed for symbiosis or in specific cells. Finally, we addressed whether differences in RNA processing might influence RNA translatability and/or stability.
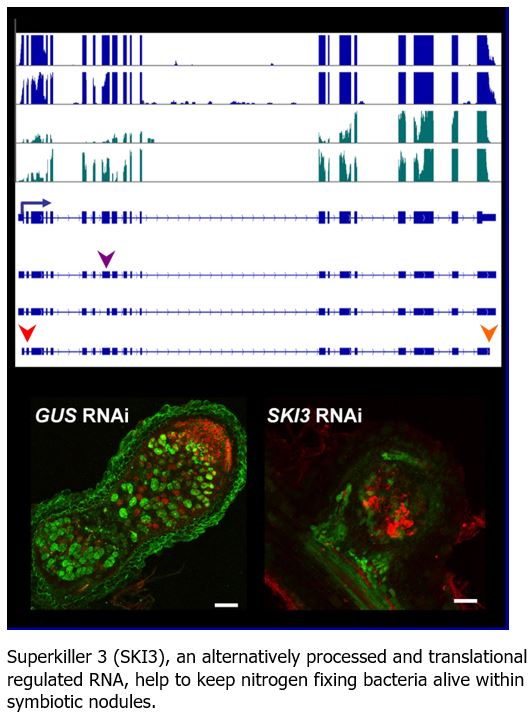
Findings: Inoculation of Medicago truncatula roots with nitrogen-fixing bacteria triggered profound changes in RNA translation, which were partially independent of changes in transcription. This regulation was also influenced by the root cell-type engaged in symbiosis. Translation dynamically adjusted the expression of regulatory genes, such as those involved in chromatin remodeling, transcription and RNA decay. Alternative processing of RNAs also influenced their translational status or response, contributing to regulate gene expression during symbiosis. This was the case for non-coding RNAs that are precursors of small regulatory RNAs, but also for an RNA that encodes the SUPERKILLER 3 protein involved in RNA degradation. SUPERKILLER 3 contributes to keep bacteria alive within nodules, and thus, to the success of the nitrogen-fixing symbiosis.
Next steps: We aim to investigate how RNA degradation mechanisms alter the repertoire of RNAs present in symbiotic cells and whether interfering with these mechanisms of RNA degradation affect the establishment of the symbiosis. Dissecting these mechanisms should help to optimize nitrogen fixation and improve crop productivity in agriculture.
Soledad Traubenik, Mauricio Alberto Reynoso, Karen Hobecker, Marcos Lancia, Maureen Hummel, Benjamin Rosen, Christopher Town, Julia Bailey-Serres, Flavio Blanco, María Eugenia Zanetti. (2019). Reprogramming of root cells during nitrogen-fixing symbiosis involves dynamic polysome association of coding and non-coding RNAs. Plant Cell. https://doi.org/10.1105/tpc.19.00647.
Key words: translation, non-coding RNAs, mRNA degradation, nodule symbiosis