Transcriptome Studies of Deepwater Rice
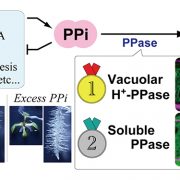
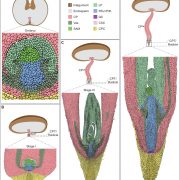
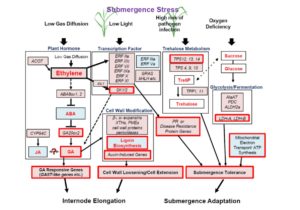 Rice (Oryza sativa) paddies frequently become submerged during the rainy season in some parts of South and Southeast Asia, such as Bangladesh, India, Thailand, Vietnam, and Cambodia. Submergence stress is harmful to plants. In addition to causing O2– and CO2-deficient conditions by restricting environmental gas exchange, submergence reduces the light available for photosynthesis, perturbs cellular energy generation and disrupts ionic balance. Deepwater rice can grow and survive in such conditions even under several month-long periods of deep flooding. Deepwater rice adapts to submergence by rapidly elongating its internodes, thereby maintaining its leaves above the water surface. To gain further insight into this adaptation, Minami et al. (10.1104/pp.17.00858) performed a comparative RNA sequencing transcriptome analysis of the shoot base region, including basal nodes, internodes and shoot apices of seedlings at two developmental stages from two rice varieties with markedly different deepwater growth responses. A transcriptomic comparison between deepwater rice C9285 and non-deepwater rice Taichung 65 revealed both similar and differential expression patterns between the two genotypes in response to submergence. The expression of genes related to gibberellin biosynthesis, trehalose biosynthesis, anaerobic fermentation, cell wall modification and transcription factors that include ethylene-responsive factors was significantly different between the varieties. Interestingly, in both varieties the jasmonic acid content at the shoot base decreased during submergence, while exogenous jasmonic acid inhibited submergence-induced internode elongation in C9285, suggesting that jasmonic acid plays a role in the submergence response of rice. Transcripts were also found that were specific to C9285, including submergence-induced biotic stress-related genes.
Rice (Oryza sativa) paddies frequently become submerged during the rainy season in some parts of South and Southeast Asia, such as Bangladesh, India, Thailand, Vietnam, and Cambodia. Submergence stress is harmful to plants. In addition to causing O2– and CO2-deficient conditions by restricting environmental gas exchange, submergence reduces the light available for photosynthesis, perturbs cellular energy generation and disrupts ionic balance. Deepwater rice can grow and survive in such conditions even under several month-long periods of deep flooding. Deepwater rice adapts to submergence by rapidly elongating its internodes, thereby maintaining its leaves above the water surface. To gain further insight into this adaptation, Minami et al. (10.1104/pp.17.00858) performed a comparative RNA sequencing transcriptome analysis of the shoot base region, including basal nodes, internodes and shoot apices of seedlings at two developmental stages from two rice varieties with markedly different deepwater growth responses. A transcriptomic comparison between deepwater rice C9285 and non-deepwater rice Taichung 65 revealed both similar and differential expression patterns between the two genotypes in response to submergence. The expression of genes related to gibberellin biosynthesis, trehalose biosynthesis, anaerobic fermentation, cell wall modification and transcription factors that include ethylene-responsive factors was significantly different between the varieties. Interestingly, in both varieties the jasmonic acid content at the shoot base decreased during submergence, while exogenous jasmonic acid inhibited submergence-induced internode elongation in C9285, suggesting that jasmonic acid plays a role in the submergence response of rice. Transcripts were also found that were specific to C9285, including submergence-induced biotic stress-related genes.