To Golgi and Beyond!
The Golgi apparatus is the central sorting station of the eukaryotic secretory pathway. Protein and lipid cargoes are received at its cis face from the endoplasmic reticulum (ER) and may undergo various modifications including glycosylation before being trafficked onwards from the trans face to their final cellular destination. In addition, the Golgi serves as a major organelle in the synthesis of cell wall polysaccharides (pectin and hemicellulose). The organelle itself consists of a series of stacked cisternae that are both structurally and functionally distinct. Such modular organization increases the efficiency of both complex molecule biosynthesis and secretion (Nakamura et al., 2012). However, little is known about the underlying spatial partitioning of Golgi-resident proteins between the cis, medial or trans cisternae, nor the key sequence characteristics that determine such intra-Golgi localization.
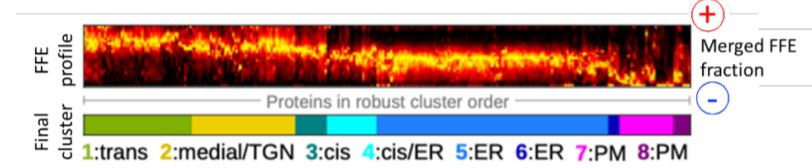
A new study by Parsons et al. (2019) exploits the innate gradient of increasingly negative surface charge that exists from the ER and across the Golgi, by employing free-flow electrophoresis (FFE) to separate Golgi sub-compartments in Arabidopsis suspension-culture cells (Islinger et al., 2010). This high-resolution fractionation was subsequently coupled with high-throughput shotgun proteomics to obtain a detailed characterization of the ER and Golgi sub-proteomes (see Figure). Robust hierarchical clustering of the identified proteins according to their FFE fractional abundance enabled their classification into eight distinct organelle-specific clusters.
 The discrete separation of the Golgi cisternae by FFE was validated by both super-resolution fluorescence imaging of selected cis-, medial– and trans-Golgi candidate proteins; as well as in situ immuno-gold labelling of glycan epitopes across the Golgi stacks. As proteins migrate through the Golgi, they become progressively glycosylated. In accordance, polysaccharides with increasing structural complexity were observed to localize to later cisternae in vivo, with the individual sub-proteomes also containing members of the corresponding biosynthetic enzyme families.
The discrete separation of the Golgi cisternae by FFE was validated by both super-resolution fluorescence imaging of selected cis-, medial– and trans-Golgi candidate proteins; as well as in situ immuno-gold labelling of glycan epitopes across the Golgi stacks. As proteins migrate through the Golgi, they become progressively glycosylated. In accordance, polysaccharides with increasing structural complexity were observed to localize to later cisternae in vivo, with the individual sub-proteomes also containing members of the corresponding biosynthetic enzyme families.
Crucially, the authors were able to distinguish between Golgi-resident proteins and secretory cargo within the sub-organelle proteome datasets. By complementing their FFE approach with density gradient centrifugation and quantitative isotope tagging, profiles of proteins localized to multiple organelles (i.e. cargo), as well as non-secretory contaminants such as chloroplast or mitochondrial proteins, were identified and ultimately excluded. The lack of any obvious cluster enrichment of such secretory cargoes also aligns with the consensus model of continuous and uniform trafficking along the cis– to trans-Golgi axis (Luini, 2011)
Mitochondrial, chloroplast and nuclear proteins carry known transit peptides at their N-terminus or nucleus localization signals, respectively, which can be recognized by machine-learning algorithms. Moreover, soluble and integral membrane proteins that are synthesized at the ER often contain N-terminal signal sequences as well as additional motifs for their proper trafficking within the endomembrane system. However, the specific sub-cellular localization of proteins resident within the secretory pathway is more difficult to predict based on in silico sequence analysis alone due to limitations in our current knowledge of such targeting motifs. Bioinformatic analyses of the transmembrane (TM) regions of proteins resident within the ER, cis-, medial– and trans-Golgi compartments and also the plasma membrane (PM) revealed characteristic sequence-specific motifs and physiochemical properties that may determine their localization. Notably, a gradient of preference for serine and phenylalanine at the luminal side of the TM, or near-TM, region existed from the ER through cis– to trans-Golgi; and the frequency of positively charged residues at the cytoplasmic TM edge increased from the ER to PM. Moreover, whilst the average length of the predicted TM domain shows a broad increase along the whole secretory pathway, this TM characteristic becomes significant from the medial Golgi onwards.
Taken together, these identified TM protein features exhibit a continuum of differences rather than discrete localization ‘rules’ per se. This is further reflected in the sub-proteomes themselves, whereby protein families with known biosynthetic functionality were localized in sub-Golgi compartments consistent with the progressive nature of cargo glycosylation and the recycling of resident proteins between adjacent compartments.
This high confidence and comprehensive dataset of ER- and Golgi-resident proteins, at sub-organelle level resolution, highlights the complexity and functional diversity of the Golgi. The challenge now is to elucidate how this pivotal organelle regulates the sorting of secreted proteins to their final destination.
REFERENCES
Islinger, M., Eckerskorn, C., and Völkl, A. (2010). Free-flow electrophoresis in the proteomic era: A technique in flux. ELECTROPHORESIS 31: 1754–1763.
Luini, A. (2011). A brief history of the cisternal progression-maturation model. Cellular Logistics 1: 6–11.
H.T. Parsons, T. J. Stevens, H. E. McFarlane, S. Vidal-Melgosa, J. Griss, N. Lawrence, R. Butler, M. M. L. Sousa, M. Salemi, W. G. T. Willats, C. J. Petzold, J. L. Heazlewood, K. S. Lilley (2019) Separating Golgi proteins from cis to trans reveals underlying properties of cisternal. Plant Cell doi: 10.1105/tpc.19.00081
Nakamura, N., Wei, J.-H., and Seemann, J. (2012). Modular organization of the mammalian Golgi apparatus. Current Opinion in Cell Biology 24: 467–474.
Figure legend:
Electrophoretic separation and clustering analysis of Arabidopsis endomembrane proteins
Membrane-bound compartments from Arabidopsis cell-suspension cultures were separated according to their surface change by free-flow electrophoresis and their proteomes determined at sub-organelle level resolution.
[Adapted from Parsons et al., (2019), Figure 4]