Tissue-resolved multi-omics atlas of Arabidopsis (Nature)
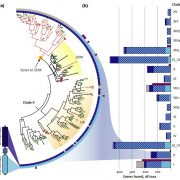
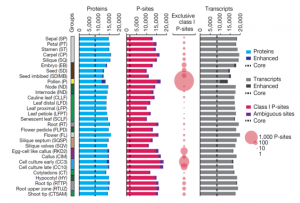 Arabidopsis as a model system has been intensively studied over the past twenty years, but the proteome of Arabidopsis has been less well characterized. Here, Mergner et al. report the first 30-tissue-type integrated proteome, phosphoproteome and transcriptome atlas of Arabidopsis. The data cover the abundance of more than 18,000 proteins out of more than 27,000 annotated protein-coding genes, and identifies nearly 44,000 phosphorylation sites (P-sites), making this one of the most extensive studies so far. In general, these findings show a positive correlation between transcript and protein level in almost all the tissue types. Mass spectrometry analysis revealed spice-site variants and peptides translated from short open-reading frames, demonstrating the power of these tools to improve genome annotation. The study detected about 700 kinases and 120 phosphatases across most of the tissues. By motif-X algorithm, this study identifies 266 phosphorylation motifs grouped into different classes, and the most complete Arabidopsis phosphoproteome to date. Overall, this study presents a deeply analysed multi-omics data set. As an example of its power, the authors show how it can reveal tissue-specific complexes or motif-based phosphorylation events. These data can be accessed by using web portals ATHENA and ProteomicsDB. (Summary by Min May Wong @wongminmay) Nature 10.1038/s41586-020-2094-2
Arabidopsis as a model system has been intensively studied over the past twenty years, but the proteome of Arabidopsis has been less well characterized. Here, Mergner et al. report the first 30-tissue-type integrated proteome, phosphoproteome and transcriptome atlas of Arabidopsis. The data cover the abundance of more than 18,000 proteins out of more than 27,000 annotated protein-coding genes, and identifies nearly 44,000 phosphorylation sites (P-sites), making this one of the most extensive studies so far. In general, these findings show a positive correlation between transcript and protein level in almost all the tissue types. Mass spectrometry analysis revealed spice-site variants and peptides translated from short open-reading frames, demonstrating the power of these tools to improve genome annotation. The study detected about 700 kinases and 120 phosphatases across most of the tissues. By motif-X algorithm, this study identifies 266 phosphorylation motifs grouped into different classes, and the most complete Arabidopsis phosphoproteome to date. Overall, this study presents a deeply analysed multi-omics data set. As an example of its power, the authors show how it can reveal tissue-specific complexes or motif-based phosphorylation events. These data can be accessed by using web portals ATHENA and ProteomicsDB. (Summary by Min May Wong @wongminmay) Nature 10.1038/s41586-020-2094-2
[altmetric doi=”10.1038/s41586-020-2094-2″ details=”right” float=”right”]