Thinking Outside the Plant: Exploring Phloem Development Using VISUAL
IN BRIEF by Jennifer Lockhart [email protected]
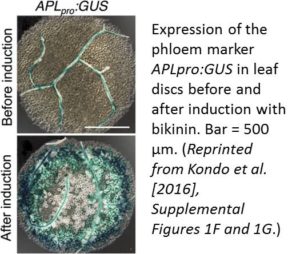 Investigating how plants grow and develop often requires a bit of creativity. For example, deep within the plant, the vascular cambium, a layer of embryonic, highly cytoplasmic cells, gives rise to xylem and phloem tissue, which must expand throughout the plant’s lifetime. Water and nutrients primarily flow through the xylem, whereas photosynthate and signaling molecules travel through the phloem. While anatomical and labeling studies have shed light on the sequential development of vascular tissue, this process is notoriously difficult to track due to the limited accessibility of the plant vasculature. However, under the proper conditions, various cells can be coaxed into producing vascular cells in culture, which greatly facilitates their investigation. Such analyses have revealed many key factors that regulate the differentiation of xylem cells from vascular cambium cells (reviewed in Růžička et al., 2015). However, unlike xylem cells, which undergo marked changes during development, in vitro-produced phloem cells are difficult to distinguish from other cell types without the use of phloem-specific markers.
Investigating how plants grow and develop often requires a bit of creativity. For example, deep within the plant, the vascular cambium, a layer of embryonic, highly cytoplasmic cells, gives rise to xylem and phloem tissue, which must expand throughout the plant’s lifetime. Water and nutrients primarily flow through the xylem, whereas photosynthate and signaling molecules travel through the phloem. While anatomical and labeling studies have shed light on the sequential development of vascular tissue, this process is notoriously difficult to track due to the limited accessibility of the plant vasculature. However, under the proper conditions, various cells can be coaxed into producing vascular cells in culture, which greatly facilitates their investigation. Such analyses have revealed many key factors that regulate the differentiation of xylem cells from vascular cambium cells (reviewed in Růžička et al., 2015). However, unlike xylem cells, which undergo marked changes during development, in vitro-produced phloem cells are difficult to distinguish from other cell types without the use of phloem-specific markers.
Kondo et al. (2016) developed a powerful system for analyzing phloem differentiation called VISUAL (Vascular Cell Induction Culture System Using Arabidopsis Leaves). In this system, mesophyll cells from Arabidopsis thaliana leaf discs or cotyledons are induced to form procambial cells, which then develop into xylem (Kondo et al., 2015) or phloem cells. To unlock the capacity of mesophyll cells to form vascular cells in vitro, including sieve element (SE)-like phloem cells and tracheary elements (xylem), the cells are treated with bikinin, an inhibitor of the kinase GSK3, which normally suppresses vascular tissue differentiation. Induction with bikinin triggers the formation of numerous phloem precursor cells and differentiating SEs, as revealed by the increased expression of the phloem markers APLpro:GUS (β-glucuronidase gene driven by the ALTERED PHLOEM DEVELOPMENT promoter; see figure) and SEOR1pro:SEOR1-YFP (yellow fluorescent protein gene driven by the SIEVE ELEMENT OCCLUSION-RELATED1 promoter), respectively. DNA inhibitor analysis indicated that cell division is required prior to phloem cell differentiation in VISUAL. In vitro and in vivo phloem cell differentiation appear to share several crucial steps, such as the formation of SE reticulum, bundles of P-protein filaments, and thick primary cell walls, as well as an enucleation-like event. The induced phloem cells mainly consist of SEs but not companion cells, as shown by microarray analysis. The authors used flow cytometry to isolate phloem SEs expressing SEOR1pro:SEOR1-YFP, finding that bikinin-induced mesophyll cells gave rise to 300 times more YFP-positive cells than uninduced mesophyll cells. In addition, phloem-specific genes were highly expressed in YFP-positive cells, whereas xylem-specific genes were highly expressed in YFP-negative cells.
The authors then took advantage of VISUAL to investigate phloem SE development in an apl mutant, which is embryo lethal, since APL is a crucial transcription factor. Microarray analysis revealed that APL preferentially upregulates many phloem-specific genes in VISUAL, especially those functioning during later SE development. Next, three different sets of VISUAL transcriptome data, including time-course, SEOR1 cell-sorting, and apl mutant data, were used to construct networks of genes related to SE-like cell differentiation. This analysis identified a transcription factor gene, NAC020, that appears to be expressed upstream of APL. Indeed, NAC020 was expressed in apl, confirming its function upstream of APL. Both overexpression and targeted repression of NAC020 led to reduced APL expression, as well as partially inhibited SE differentiation. These findings point to the importance of NAC020 in early phloem development, and they hint at the great potential of using VISUAL to explore phloem development outside the plant.
REFERENCES
Kondo, Y., Fujita, T., Sugiyama, M., Fukuda, H. (2015). A novel system for xylem cell differentiation in Arabidopsis thaliana. Mol. Plant 8: 612–621.
Kondo, Y., Nurani, A.M., Saito, C., Ichihashi, Y., Saito, M., Yamazaki, K., Mitsuda, N., Ohme-Takagi, M., Fukuda, H. (2016). Vascular cell induction culture system using Arabidopsis leaves (VISUAL) reveals the sequential differentiation of sieve element-like cells. Plant Cell 28: 1250–1262.
Růžička, K., Ursache, R., Hejátko, J., Helariutta, Y. (2015). Xylem development – from the cradle to the grave. New Phytol. 207: 519–535.






Leave a Reply
Want to join the discussion?Feel free to contribute!