The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor (Nature Gen.)
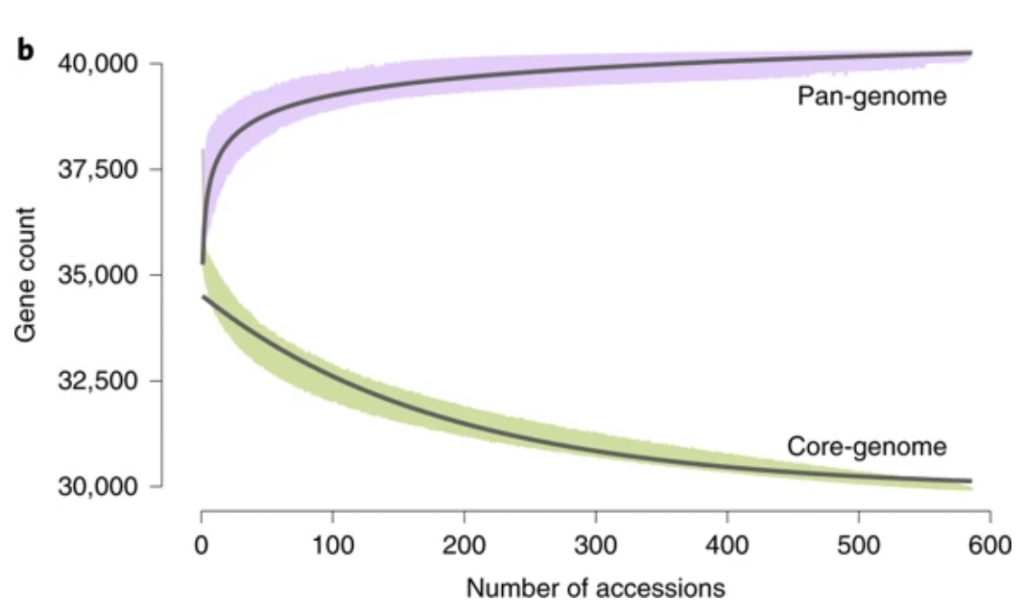
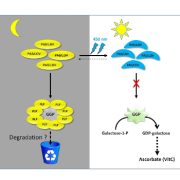
The wide variety of shape and flavor in modern cultivated tomatoes is a result of introgressions from its wild relatives. While many genetic studies were successful in the identification of wild alleles that affect tomato aroma, no tools were available to study structural variants affecting the taste, missing from the reference line. Gao et al. used the genomic sequences of large-fruit sized tomatoes, cherry tomatoes, and tomato’s closest wild relatives, and by de novo assembly was able to identify previously unknown sequences, not present in the reference genome. A total of 5,000 protein-coding genes were predicted in the sequences not represented in the reference genome. Additionally, almost 26% of genes in the pan-genome showed varying degree of presence/absence variation, indicating the diverse makeup of Lycopersidae clade. The wild tomato genomes were not only containing more genes, but they were enriched in defense responses, oxidation-reduction, and senescence. The pan-genome analysis enabled to identify a 4 kb substitution in the promoter region of TomLoxC, which was also mapped using a QTL study to be responsible for the variation in for volatiles and apocarotenoids production. (Summarized by Magdalena Julkowska) Nature Genetics 10.1038/s41588-019-0410-2