The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion ($)
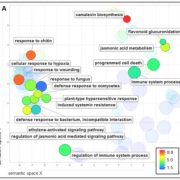
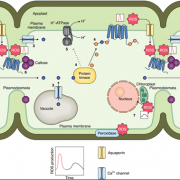
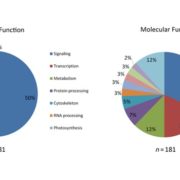
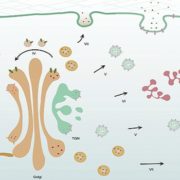
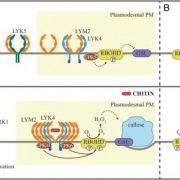
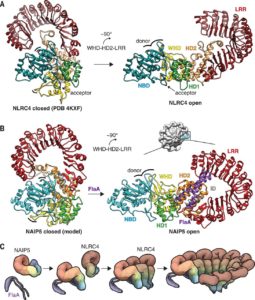 Both plants and animals have a sophisticated immune system comprising receptors to detect pathogen-encoded epitopes or virulence molecules. These receptors are programmed to recognize pathogen ligands which are rapidly evolving over an evolutionary time. However, the structural basis of ligand perception by immune receptors that encode nucleotide-binding domain leucine-rich repeat (NLR) proteins and of their further activation is still unknown. Tenthorey et al. have shed light onto this process by structurally dissecting one of the best characterized NLRs in mouse NAIP5. NAIPs co-oligomerise with NLRC4 and form a high molecular weight structure called the inflammasome. By using the Cryo-EM technology, the authors resolve the ~1.4 megadalton structural details of NAIP5-NLRC4 inflammasome bound to flagellin from Legionella. These set of experiments beautifully reveal the mechanism of NLR activation by a ligand. Upon binding to flagellin, NAIP5 undergoes a conformational change triggering rotation of monomeric NLRC4, thereby disrupting the inflammasome structure. Multiple flagellin surfaces are recognized by NAIP5 and multiple mutations on flagellin to evade NAIP5 recognition has a huge fitness cost for the pathogen. This provides a solid structural and functional evidence of multisurface recognition in immune activation to withhold the pathogen immune escape. (Summary by Amey Redkar) Science 10.1126/science.aao1140
Both plants and animals have a sophisticated immune system comprising receptors to detect pathogen-encoded epitopes or virulence molecules. These receptors are programmed to recognize pathogen ligands which are rapidly evolving over an evolutionary time. However, the structural basis of ligand perception by immune receptors that encode nucleotide-binding domain leucine-rich repeat (NLR) proteins and of their further activation is still unknown. Tenthorey et al. have shed light onto this process by structurally dissecting one of the best characterized NLRs in mouse NAIP5. NAIPs co-oligomerise with NLRC4 and form a high molecular weight structure called the inflammasome. By using the Cryo-EM technology, the authors resolve the ~1.4 megadalton structural details of NAIP5-NLRC4 inflammasome bound to flagellin from Legionella. These set of experiments beautifully reveal the mechanism of NLR activation by a ligand. Upon binding to flagellin, NAIP5 undergoes a conformational change triggering rotation of monomeric NLRC4, thereby disrupting the inflammasome structure. Multiple flagellin surfaces are recognized by NAIP5 and multiple mutations on flagellin to evade NAIP5 recognition has a huge fitness cost for the pathogen. This provides a solid structural and functional evidence of multisurface recognition in immune activation to withhold the pathogen immune escape. (Summary by Amey Redkar) Science 10.1126/science.aao1140