The prevalence, evolution and chromatin signatures of plant regulatory elements ($) (Nature Plants)
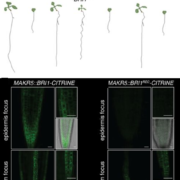
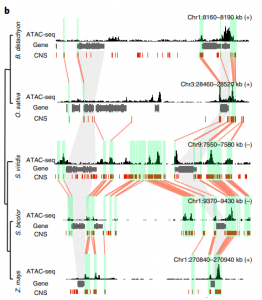 With a wide range of genome sizes and varying architectures, understanding the evolution and function of non-coding regions is not straightforward in plants. Nevertheless, many advances are coming out that will shed light on this complex scientific problem. Lu et al. performed ATAC-seq to study chromatin accessibility in 13 plant species spanning most angiosperm taxonomic groups. They found that accessible chromatin region (ACR) distribution correlates with genome size and their localization relative to transcription starting sites is very different among species. In many cases, ACRs are conserved across species, in terms of sequence and relative position to genes, even for distal ACRs (>2kb). Interestingly, transposable elements played a major role in the evolution of ACR distribution among plant genomes. They also studied chromatin features in all analyzed species and made them available online for researchers (http://epigenome.genetics.uga.edu/PlantEpigenome/). Accessible regions also possess characteristic histone modifications similarly to animal genomes but lacking H3K4me1. These chromatin states in ACRs are also conserved in different species suggesting they could be important for gene regulation. Future works will help to validate their function in planta. (Summary by Facundo Romani) Nature Plants 10.1038/s41477-019-0548-z
With a wide range of genome sizes and varying architectures, understanding the evolution and function of non-coding regions is not straightforward in plants. Nevertheless, many advances are coming out that will shed light on this complex scientific problem. Lu et al. performed ATAC-seq to study chromatin accessibility in 13 plant species spanning most angiosperm taxonomic groups. They found that accessible chromatin region (ACR) distribution correlates with genome size and their localization relative to transcription starting sites is very different among species. In many cases, ACRs are conserved across species, in terms of sequence and relative position to genes, even for distal ACRs (>2kb). Interestingly, transposable elements played a major role in the evolution of ACR distribution among plant genomes. They also studied chromatin features in all analyzed species and made them available online for researchers (http://epigenome.genetics.uga.edu/PlantEpigenome/). Accessible regions also possess characteristic histone modifications similarly to animal genomes but lacking H3K4me1. These chromatin states in ACRs are also conserved in different species suggesting they could be important for gene regulation. Future works will help to validate their function in planta. (Summary by Facundo Romani) Nature Plants 10.1038/s41477-019-0548-z