The pentatricopeptide repeat PPR-E proteins mediate RNA C-to-U editing by recruiting a cytidine deaminase
Wang et al. reveal how the PPR-E proteins function in RNA editing in maize mitochondria.
Yong Wang and Bao-Cai Tan
Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China
Background: Mitochondria produce energy for all cell activities and contain genomes harboring about 60 genes. Mitochondrial gene transcripts undergo complex post-transcriptional modifications, including RNA C-to-U editing, intron splicing, maturation of transcript ends, and RNA stabilization. RNA C-to-U editing is widespread in vascular plants and plays an essential role in organellar gene expression and plant growth and development. However, the underlying mechanism of RNA editing is not fully understood. PLS-class pentatricopeptide repeat (PPR) proteins are divided into three subclasses, PPR-E, PPR-E+, and PPR-DYW, and function as critical factors in C-to-U editing of RNAs. The DYW domain in PPR-DYW proteins provides the cytidine deaminase activity for RNA C-to-U editing and PPR-E+ proteins recruit an atypical PPR-DYW protein to function in RNA editing.
Question: The PPR-E proteins lack the E+ and DYW domains but function in RNA editing. How do PPR-E subclass proteins mediate RNA C-to-U editing?
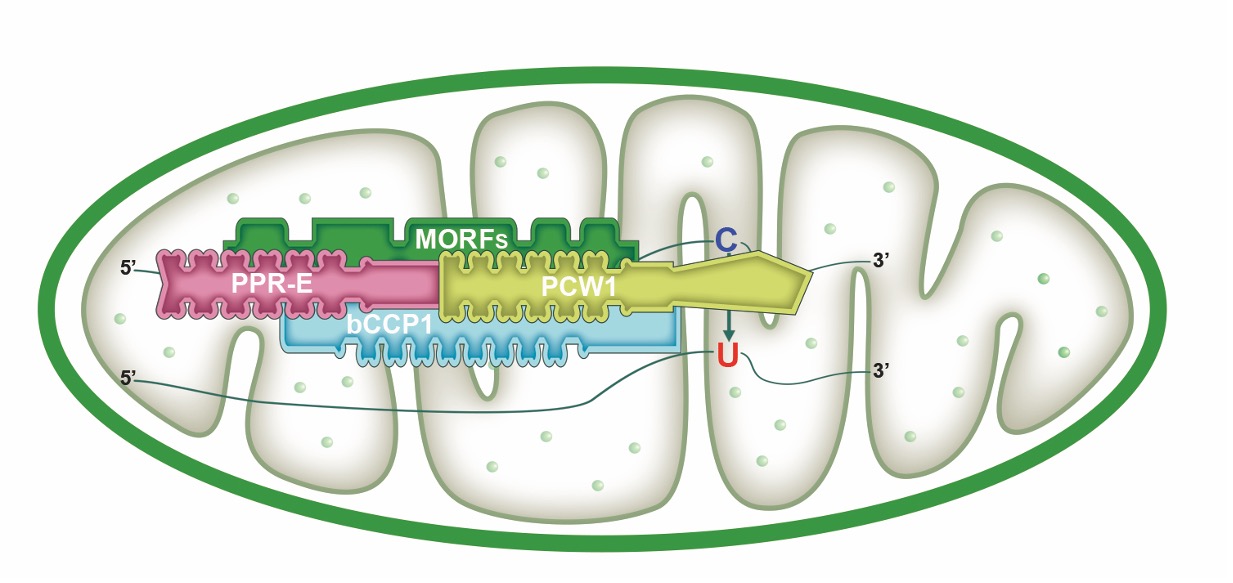 Findings: We identified bZIP and coiled-coil domain-containing PPR 1 (bCCP1) and an atypical PPR-DYW protein, PCW1, which are required for the C-to-U editing at 66 sites and 102 sites in maize (Zea mays) mitochondria, respectively. Surprisingly, the 66 editing sites overlap entirely with the 102 editing sites and are solely associated with known PPR-E class proteins. Furthermore, protein interaction analyses show that bCCP1, PCW1, and the multiple organellar RNA editing factor (MORF) proteins ZmMORF1/8 interact with each other, and the PPR-E protein Empty pericarp7 interacts with bCCP1 and ZmMORF1/8. Thus, we propose a model for PPR-E protein function in RNA editing in which the PPR-E protein recognizes the target site and recruits the trans cytidine deaminase PCW1 with the assistance of bCCP1 and MORFs.
Findings: We identified bZIP and coiled-coil domain-containing PPR 1 (bCCP1) and an atypical PPR-DYW protein, PCW1, which are required for the C-to-U editing at 66 sites and 102 sites in maize (Zea mays) mitochondria, respectively. Surprisingly, the 66 editing sites overlap entirely with the 102 editing sites and are solely associated with known PPR-E class proteins. Furthermore, protein interaction analyses show that bCCP1, PCW1, and the multiple organellar RNA editing factor (MORF) proteins ZmMORF1/8 interact with each other, and the PPR-E protein Empty pericarp7 interacts with bCCP1 and ZmMORF1/8. Thus, we propose a model for PPR-E protein function in RNA editing in which the PPR-E protein recognizes the target site and recruits the trans cytidine deaminase PCW1 with the assistance of bCCP1 and MORFs.
Next steps: The editing at 102 sites requires PCW1, but only 66 of these sites require bCCP1; therefore, one or more bCCP1-like proteins are predicted to function as a bridge between the PPR-E proteins and PCW1. What is the other bCCP1-like protein? Are there other non-PPR editing factors in the PPR-E complexes? What is the role of the non-PPR proteins in the editosome?
Reference:
Yong Wang, Hao Li, Zi-Qin Huang, Bing Ma, Yan-Zhuo Yang, Zhi-Hui Xiu, Le Wang, Bao-Cai Tan (2022) Maize PPR-E proteins mediate RNA C-to-U editing in mitochondria by recruiting the trans deaminase PCW1. Plant Cell. https://doi.org/10.1093/plcell/koac298