The miRNome function transitions from regulating developmental genes to transposable elements during pollen maturation (Plant Cell)
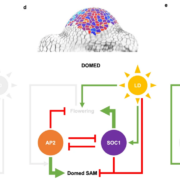
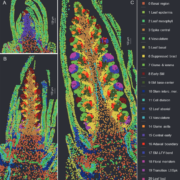
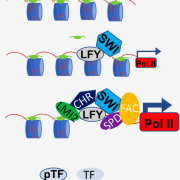
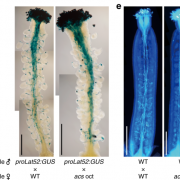
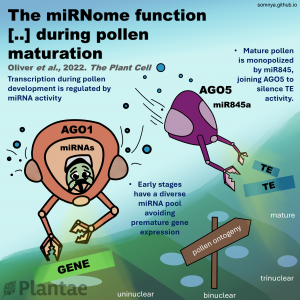 Genes (protein-coding or noncoding) and transposable elements (TEs) are the main characters in the genomic landscape, but there are few reported systems where we have seen all acting together to produce a phenotype. Oliver and collaborators analyzed miRNA populations during pollen maturation in an outstanding experimental and computational initiative. In Arabidopsis, pollen development goes from uninuclear to binuclear, trinuclear, and finally, mature pollen. Initially, the pool of miRNAs is diverse at the uninuclear stage but it becomes dominated by miR845 at the mature stage. Different miRNAs are selectively introduced to Argonaute (AGO) proteins depending on their structure and which target they pursue. AGO1 is commonly present in the uninuclear stage, repressing coding transcripts related to molecular functions. As pollen matures, AGO5 appears and associates with miR845a to cleave TEs. Jointly, miRNAs and AGO proteins build a bivalent system that starts in gene regulation and ends in TE post-transcriptional silencing and, presumably, siRNA production. Many exciting questions arise from this work, such as is it a conserved ontogenetic system in plants? Could plant development be predicted by looking at the genomic and transcriptomic landscape? This unique work should be introduced into each plant physiology and genomics course. (Summary and sketch by Eddy Mendoza-Galindo @IamSomnya) Plant Cell 10.1093/plcell/koab280
Genes (protein-coding or noncoding) and transposable elements (TEs) are the main characters in the genomic landscape, but there are few reported systems where we have seen all acting together to produce a phenotype. Oliver and collaborators analyzed miRNA populations during pollen maturation in an outstanding experimental and computational initiative. In Arabidopsis, pollen development goes from uninuclear to binuclear, trinuclear, and finally, mature pollen. Initially, the pool of miRNAs is diverse at the uninuclear stage but it becomes dominated by miR845 at the mature stage. Different miRNAs are selectively introduced to Argonaute (AGO) proteins depending on their structure and which target they pursue. AGO1 is commonly present in the uninuclear stage, repressing coding transcripts related to molecular functions. As pollen matures, AGO5 appears and associates with miR845a to cleave TEs. Jointly, miRNAs and AGO proteins build a bivalent system that starts in gene regulation and ends in TE post-transcriptional silencing and, presumably, siRNA production. Many exciting questions arise from this work, such as is it a conserved ontogenetic system in plants? Could plant development be predicted by looking at the genomic and transcriptomic landscape? This unique work should be introduced into each plant physiology and genomics course. (Summary and sketch by Eddy Mendoza-Galindo @IamSomnya) Plant Cell 10.1093/plcell/koab280