The Lipase Link: Abscisic Acid Induces PLASTID LIPASES, Which Produce Jasmonic Acid Precursors
Crosstalk, crosstalk— it’s a word that keeps coming up. Indeed, and perhaps not surprisingly, plant hormone signaling pathways all seem to affect each other to some extent. For example, the MYC2 transcription factor plays roles in abscisic acid (ABA) and jasmonic acid (JA) signaling in the response to abiotic stress. Moreover, ABA stimulates JA biosynthesis; however, the mechanism for this remains unclear. Now, Wang et al. (2018), have found enzymes that play a key role in the crosstalk between ABA and JA. The authors come at the question via examination of lipid breakdown in chloroplast membranes. These crucial membranes maintain their complex, unique lipid composition by balancing lipid biosynthesis and breakdown (reviewed in Kelly and Feussner, 2016).
For lipid breakdown, the Arabidopsis thaliana genome encodes hundreds of predicted lipases; different lipases cleave at different positions on the lipid and release fatty acids that can be oxygenated and converted into other compounds, including JA, by various enzymes. The authors had previously identified PLASTID LIPASE1 (PLIP1), which localizes to the chloroplast and participates in seed oil production (Wang et al., 2017). Here, they extend their studies to the PLIP1 homologs PLIP2 and PLIP3. Chloroplast import assays and examination of fluorescent protein fusions showed that (like PLIP1) PLIP2 and PLIP3 localize to the chloroplast, albeit in different subplastid compartments (PLIP1, thylakoids; PLIP2, envelope membranes, stroma, thylakoids; PLIP3, envelope membranes, thylakoids).
Galactolipids, that is lipids containing the sugar galactose instead of the phosphate found in phospholipids, make up a large part of the chloroplast membrane. Chloroplast lipids include, among others, monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and the anionic lipid phosphatidylglycerol (PG). The authors previously showed that PLIP1 functions as a lipase (type A1 hydrolyses lipids at the sn-1 position) and they further conduced in vitro lipase assays, and lipidome profiling and in vivo pulse-chase experiments in PLIP-overexpressing (PLIP-OX) plants. These assays showed that PLIP2 and PLIP3 have glycerolipase A1 activity, with PLIP2 showing a preference for MGDG and PLIP3 having a preference for PG.
Intriguingly, the PLIP2-OX and PLIP3-OX plants showed severely stunted growth (more severe than PLIP1-OX), short petioles, and pigment accumulation (presumably anthocyanin), similar to plants with activated JA signaling. Indeed, the authors found that JA-related genes were upregulated in PLIP2-OX and PLIP3-OX plants, including genes related to JA biosynthesis, catabolism, signaling (including MYC2), and negative regulation. Moreover, targeted quantification and non-targeted metabolite profiling showed that the PLIP2-OX and PLIP3-OX plants accumulated high levels of JA compounds and the related Arabidopside galactolipids. The activation of JA signaling appears to cause the stunted phenotype, as PLIP2-OX and PLIP3-OX showed normal growth in the coronatine insensitive 1 (coi1) mutant background, which lacks a component of the JA receptor complex.
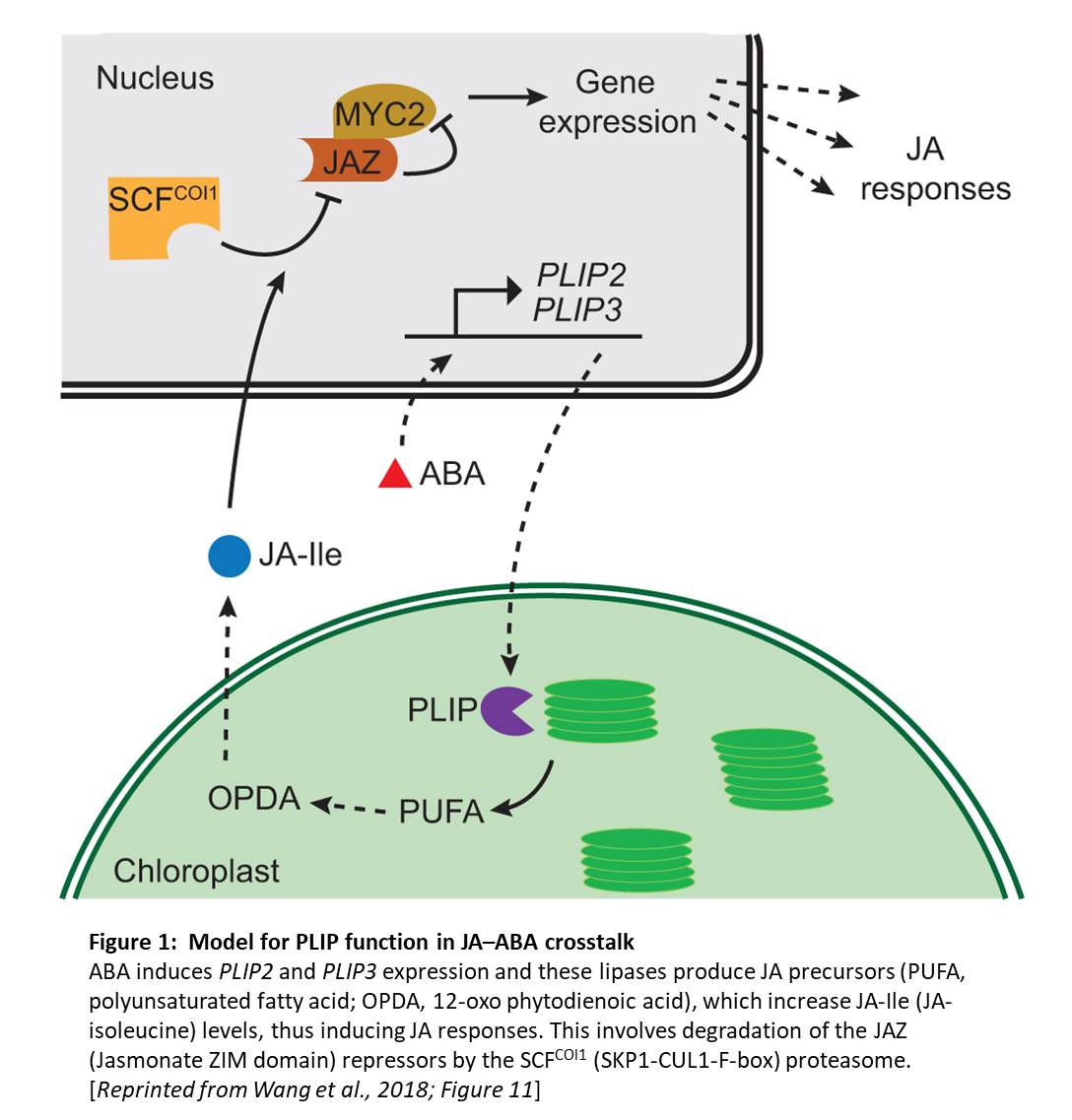
Given the strong effects of PLIP overexpression, the authors next examined PLIP2 and PLIP3 expression. To this end, they queried Arabidopsis transcriptomic data and found that PLIP expression occurs in some specific tissues (PLIP2 in pollen and PLIP3 in pollen, embryos, and senescent tissue), and that ABA and abiotic stress strongly induced PLIP2 and PLIP3 expression. The authors produced plip triple mutants and found that these mutants failed to produce more JA in response to ABA; moreover, germination and growth assays showed that the plip triple mutants were hypersensitive to ABA. Therefore, this study provides a long-awaited mechanistic explanation for the known crosstalk between ABA and JA signaling pathways (see figure). Examining the different roles of these three PLIPs in ABA- and JA-related plant developmental processes, and in the responses to specific stresses, remains an interesting prospect for future research.
REFERENCES
Kelly, A.A., and Feussner, I. (2016). Oil is on the agenda: Lipid turnover in higher plants. Biochim Biophys Acta. 1861, 1253–1268
Wang, K., Froehlich, J.E., Zienkiewicz, A., Hersh, H.L., and Benning, C. (2017). A Plastid Phosphatidylglycerol Lipase Contributes to the Export of Acyl Groups from Plastids for Seed Oil Biosynthesis. Plant Cell 29, 1678-1696.
Wang, K., Guo, Q., Froehlich, J.E., Hersh, H. L., Zienkiewicz, A., Howe, G.A., Benning, C. (2018). Two Abscisic Acid Responsive Plastid Lipase Genes Involved in Jasmonic Acid Biosynthesis in Arabidopsis thaliana. Plant Cell DOI: https://doi.org/10.1105/tpc.18.00250.