The highly buffered Arabidopsis immune signaling network conceals the functions of its components
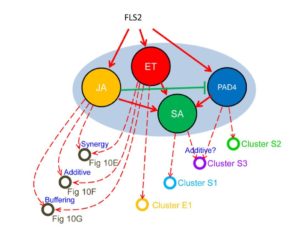 Classic genetic approaches have been instrumental in identifying genes that control developmental and defense networks, but as Hillmer et al. point out, analysis of single mutants is complicated by network buffering. They describe network buffering as occurring when the effect of losing a gene’s function is masked by the function of other genes. They point out that the immune signaling network must be highly buffered as it is constantly under attack by pathogens. Previously, they defined the immune network as being made up of four sub-networks, each of which can be interrupted by disabling a single gene. Specifically, they can eliminate the jasmonate, salicylate, ethylene and PAD4 sectors by elimination of a biosynthetic or signaling gene. They previously published an analysis of higher order mutants knocking out all combinations of these sub-networks. Here they analyze transcriptomic and hormone profiles of each of these mutants in response to flg22, a peptide that mimics the presence of a pathogen. Their results demonstrate that the immune network “is not a collection of independent pathways; rather interactions among the sectors dominate network regulation of the transcriptome response.” PLOS Genet. 10.1371/journal.pgen.1006639
Classic genetic approaches have been instrumental in identifying genes that control developmental and defense networks, but as Hillmer et al. point out, analysis of single mutants is complicated by network buffering. They describe network buffering as occurring when the effect of losing a gene’s function is masked by the function of other genes. They point out that the immune signaling network must be highly buffered as it is constantly under attack by pathogens. Previously, they defined the immune network as being made up of four sub-networks, each of which can be interrupted by disabling a single gene. Specifically, they can eliminate the jasmonate, salicylate, ethylene and PAD4 sectors by elimination of a biosynthetic or signaling gene. They previously published an analysis of higher order mutants knocking out all combinations of these sub-networks. Here they analyze transcriptomic and hormone profiles of each of these mutants in response to flg22, a peptide that mimics the presence of a pathogen. Their results demonstrate that the immune network “is not a collection of independent pathways; rather interactions among the sectors dominate network regulation of the transcriptome response.” PLOS Genet. 10.1371/journal.pgen.1006639







Leave a Reply
Want to join the discussion?Feel free to contribute!