The Great Escape: How a Plant DNA Virus Hijacks an Imprinted Host Gene to Avoid Silencing
Plant viruses are a major threat to global food security. The large family of geminiviruses has a broad pathogenic impact on economically important crops such as maize, tomato, and cassava and causes millions of tons of losses per year worldwide (Rojas et al., 2005). Geminiviruses possess a circular DNA genome encoding proteins necessary for viral replication and virion assembly. The plant hosts have developed antiviral defense systems such as RNA interference that ensures viral post-transcriptional gene silencing (PTGS) by targeting viral transcripts, and small RNA-mediated DNA methylation that triggers transcriptional gene silencing (TGS) of the viral DNA. In an evolutionary arms race, geminiviruses have developed counter-defense systems to overcome both TGS and PTGS through multiple viral suppressors of silencing, including Rep and C2 that are produced early during viral infection and are thus instrumental for successful viral amplification (Zhang et al., 2011; Rodríguez-Negrete et al., 2013). While the anti-silencing activity of C2 itself is well described, little is known about how C2 allows the virus to escape silencing soon after infection.
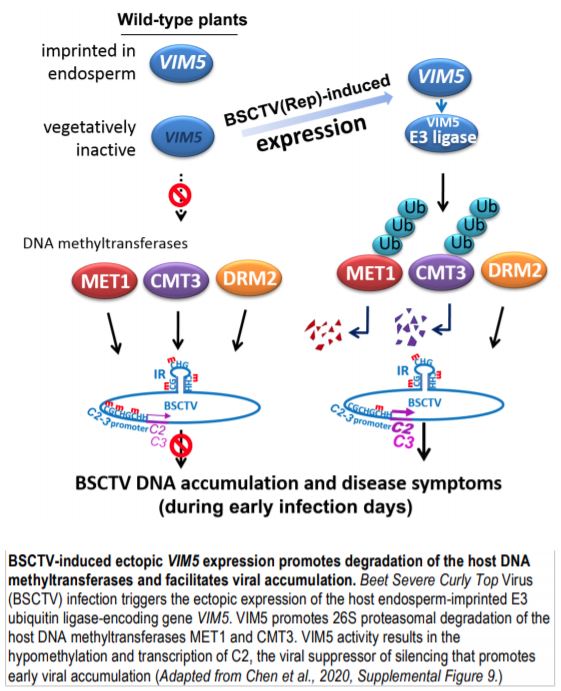 Chen et al. (2020) used Arabidopsis thaliana plants infected by the geminivirus Beet Severe Curly Top Virus (BSCTV) as a model to investigate the molecular mechanisms permitting early C2 expression. Using transgenic Arabidopsis lines carrying a BSCTV Rep transgene that also contained the C2 N-terminal sequence (C2N) under the control of the C2 promoter, the authors observed accumulation of Rep mRNA in all lines tested, but unexpectedly detected C2N transcripts in one of the transgenic lines. The accumulation of C2N transcripts correlated with hypomethylation at the C2N promoter that may have allowed transcription. To illuminate the molecular determinants of C2N transcript accumulation, the authors compared the transcriptomes of the transgenic line expressing C2N to wild-type plants and observed ectopic transcription of the imprinted E3 ubiquitin ligase-encoding gene VARIANT IN METHYLATION5 (VIM5). Overexpression of VIM5 in another transgenic line that did not show C2N transcription triggered C2N transcript accumulation and correlated with decreased DNA methylation at the transgenic C2 promoter. BSCTV infection of wild-type Arabidopsis plants recapitulated the molecular patterns observed in the transgenic plants. In BSCTV-infected vim5 mutants, the C2 promoter appeared heavily methylated and viral replication was delayed. A p35S-VIM5 transgene complemented the vim5 mutation, reduced DNA methylation at the C2 promoter, and restored early viral replication. The authors concluded that upon BSCTV infection, ectopic activation of VIM5 facilitates transcription of the viral C2 gene by limiting DNA methylation at the C2 promoter.
Chen et al. (2020) used Arabidopsis thaliana plants infected by the geminivirus Beet Severe Curly Top Virus (BSCTV) as a model to investigate the molecular mechanisms permitting early C2 expression. Using transgenic Arabidopsis lines carrying a BSCTV Rep transgene that also contained the C2 N-terminal sequence (C2N) under the control of the C2 promoter, the authors observed accumulation of Rep mRNA in all lines tested, but unexpectedly detected C2N transcripts in one of the transgenic lines. The accumulation of C2N transcripts correlated with hypomethylation at the C2N promoter that may have allowed transcription. To illuminate the molecular determinants of C2N transcript accumulation, the authors compared the transcriptomes of the transgenic line expressing C2N to wild-type plants and observed ectopic transcription of the imprinted E3 ubiquitin ligase-encoding gene VARIANT IN METHYLATION5 (VIM5). Overexpression of VIM5 in another transgenic line that did not show C2N transcription triggered C2N transcript accumulation and correlated with decreased DNA methylation at the transgenic C2 promoter. BSCTV infection of wild-type Arabidopsis plants recapitulated the molecular patterns observed in the transgenic plants. In BSCTV-infected vim5 mutants, the C2 promoter appeared heavily methylated and viral replication was delayed. A p35S-VIM5 transgene complemented the vim5 mutation, reduced DNA methylation at the C2 promoter, and restored early viral replication. The authors concluded that upon BSCTV infection, ectopic activation of VIM5 facilitates transcription of the viral C2 gene by limiting DNA methylation at the C2 promoter.
VIM5 has E3 ubiquitin ligase activity, suggesting that VIM5 maintains hypomethylation at the C2 promoter by targeting host DNA methyltransferases for degradation by the ubiquitin 26S proteasome proteolytic pathway. VIM5 indeed interacted in vitro and in vivo with the CG methyltransferase MET1 and the CHG methyltransferase CMT3, but not with the CHH methyltransferase DRM2. Co-infiltration experiments demonstrated that VIM5-dependent ubiquitination of MET1 and CMT3 proteins triggered their proteasomal degradation. To assess the molecular consequences of DNA methylation at the C2 promoter during BSCTV viral accumulation, the authors compared BSCTV infection between wild-type Arabidopsis plants and met1 and cmt3 mutants, in which CG and CHG methylation, respectively, are impaired. In agreement with previous reports, they showed that loss of MET1 and CMT3 facilitate viral replication. Accordingly, met1 and cmt3 plants exhibit lower levels of DNA methylation at the C2 viral promoter than vim5 plants. The authors generated a modified BSCTV containing 13 nucleotide substitutions at CG and CHG sites in the C2 promoter, preventing MET1- and CMT3-dependent methylation. In the vim5 background, the modified BSCTV showed higher viral accumulation compared to the unmodified BSCTV, demonstrating that DNA methylation at the viral C2 promoter inhibits viral accumulation and disease development.
While the mechanism by which BSCTV infection activates VIM5 remains an open question, this study highlights another layer of geminiviral counter-defenses through post-translational interference with the host DNA methylation machinery. The resulting impaired methylation likely threatens the genome stability of the plant host, but the molecular consequences remain to be addressed.
Matthias Benoit
Howard Hughes Medical Institute
Cold Spring Harbor Laboratory
ORCID: 0000-0002-3958-3173
REFERENCES
Rodríguez-Negrete, E., Lozano-Durán, R., Piedra-Aguilera, A., Cruzado, L., Bejarano, E.R., and Castillo, A.G. (2013). Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 199: 464–475.
Rojas, M.R., Hagen, C., Lucas, W.J., and Gilbertson, R.L. (2005). Exploiting Chinks in the Plant’s Armor: Evolution and Emergence of Geminiviruses. Annu. Rev. Phytopathol. 43: 361–394.
Zhang, Z. et al. (2011). BSCTV C2 Attenuates the Degradation of SAMDC1 to Suppress DNA Methylation-Mediated Gene Silencing in Arabidopsis. Plant Cell 23: 273–288.