The genome history of camelina emerges from the shadows
Mandáková et. al. explore the origin of an ancient oilseed crop. https://doi.org/10.1105/tpc.19.00366
Background: Camelina (Camelina sativa; gold of pleasure or false flax) is an ancient oilseed crop grown in Europe as early as 4000 BC. Gold of pleasure was largely forgotten as a crop, being replaced by oilseed rape, and recently re-discovered for its unique characteristics. The high oil content (28–40%) makes camelina an important aviation biofuel, while its omega‐3 fatty acids (“fish oils”) are beneficial for cardiovascular health. Sequencing of the camelina genome revealed that the genome consists of three parental subgenomes. However, how the hexaploid camelina genome originated and how the three subgenomes are related to other genomes in the genus Camelina remained unknown. We tackled these questions by using comparative chromosome painting, genomic in situ hybridization, and multi-gene phylogenetic analyses.
Question: We knew that the camelina genome consists of three subgenomes merged through a hybridization between older parental species. We wanted to find the most probable parental genomes of the hexaploid camelina and moreover to understand how its genome is related to genomes of other Camelina species.
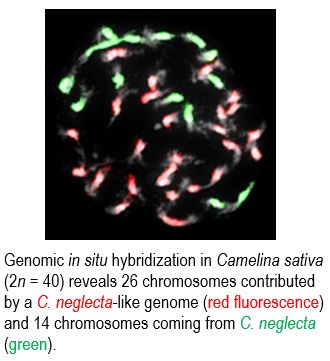 Findings: Genomes of diploid camelinas (C. hispida, n = 7 chromosomes; C. laxa, n = 6; and C. neglecta, n = 6) originated from an ancestral n = 7 genome. The allotetraploid C. rumelica genome (n = 13, N6H genome) arose from hybridization between diploids C. neglecta (n = 6, N6) and C. hispida (n = 7, H), and the N subgenome has been substantially reshuffled by chromosomal rearrangements. The allohexaploid genomes of C. sativa (n = 20, N6N7H) originated through hybridization between an auto-allotetraploid C. neglecta-like genome
Findings: Genomes of diploid camelinas (C. hispida, n = 7 chromosomes; C. laxa, n = 6; and C. neglecta, n = 6) originated from an ancestral n = 7 genome. The allotetraploid C. rumelica genome (n = 13, N6H genome) arose from hybridization between diploids C. neglecta (n = 6, N6) and C. hispida (n = 7, H), and the N subgenome has been substantially reshuffled by chromosomal rearrangements. The allohexaploid genomes of C. sativa (n = 20, N6N7H) originated through hybridization between an auto-allotetraploid C. neglecta-like genome
(n = 13, N6N7) and C. hispida (n = 7, H), and the three subgenomes remained overall stable since the genome merger. Remarkably, the ancestral and diploid Camelina genomes were shaped by complex chromosome shattering, resembling similar events associated with human disorders.
Next steps: There are multiple implications of our work. Breeders want to improve the seed yield, oil content and other agronomic traits of the camelina crop. This can be done by producing artificial camelina lines by crossing the now identified parental Camelina species. The diploid and tetraploid Camelina genomes should be more amenable to genome sequencing and uncovering the key metabolic pathways. Future work should focus on elucidating genomic homogeneity of Camelina species analyzed in our study.
Key words: camelina, genome evolution, hybridization, polyploidy




