The Functions of Retinoblastoma-Related Proteins in Rice
RETINOBLASTOMA was originally identified as a tumor suppressor gene in animals. A basic and core function of its protein is to control cell proliferation via regulating cell cycle entry. RB protein is also involved in regulating cell differentiation and organ specification. In 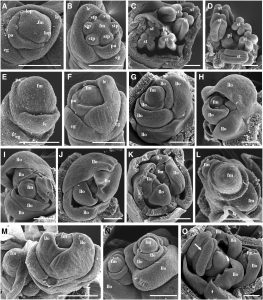 plants, RBR (RB-Related) genes are widely distributed. The mechanism of controlling the cell cycle is also highly conserved in RBR genes. However, plants possess a set of special cellular structures and developmental patterns that are distinguished from those of animals, such as the cell wall, cell totipotency, postembryonic organ formation, and the development of stem cells and meristem from a specific narrow area. These characteristics of plants may reflect differences between plant RBR and animal RB in regard to spatiotemporal expression pattern and functional mechanism as well as specific molecular interactions and responses. Duan et al. (10.1104/pp.19.00478) have characterized the functions of OsRBR1 and OsRBR2 in rice (Oryza sativa) growth and development using both forward- and reverse-genetics methods. The two genes were coexpressed and performed redundant roles in vegetative organs but exhibited separate functions in flowers. OsRBR1 was highly expressed in the floral meristem and regulated the expression of floral homeotic genes to ensure floral organ formation. Mutation of OsRBR1 caused loss of floral meristem identity, resulting in diverse floral abnormalities. OsRBR2 was preferentially expressed in stamens and promoted pollen formation. Mutation of OsRBR2 led to deformed anthers without pollen. OsRBR1 and OsRBR2 physically associated with OsMSI 1 (Multicopy Suppressors of the Iral), suggesting that their functions might be dependent on interaction with OsMSI1 at the protein level. This work further elucidates the functions of RBRs and improves our current understanding of specific regulatory pathways of floral specification and pollen formation in rice.
plants, RBR (RB-Related) genes are widely distributed. The mechanism of controlling the cell cycle is also highly conserved in RBR genes. However, plants possess a set of special cellular structures and developmental patterns that are distinguished from those of animals, such as the cell wall, cell totipotency, postembryonic organ formation, and the development of stem cells and meristem from a specific narrow area. These characteristics of plants may reflect differences between plant RBR and animal RB in regard to spatiotemporal expression pattern and functional mechanism as well as specific molecular interactions and responses. Duan et al. (10.1104/pp.19.00478) have characterized the functions of OsRBR1 and OsRBR2 in rice (Oryza sativa) growth and development using both forward- and reverse-genetics methods. The two genes were coexpressed and performed redundant roles in vegetative organs but exhibited separate functions in flowers. OsRBR1 was highly expressed in the floral meristem and regulated the expression of floral homeotic genes to ensure floral organ formation. Mutation of OsRBR1 caused loss of floral meristem identity, resulting in diverse floral abnormalities. OsRBR2 was preferentially expressed in stamens and promoted pollen formation. Mutation of OsRBR2 led to deformed anthers without pollen. OsRBR1 and OsRBR2 physically associated with OsMSI 1 (Multicopy Suppressors of the Iral), suggesting that their functions might be dependent on interaction with OsMSI1 at the protein level. This work further elucidates the functions of RBRs and improves our current understanding of specific regulatory pathways of floral specification and pollen formation in rice.



