The Case of Virus-Induced Plant Autophagy: Cui Bono?
Plant-viral interactions, like other plant-pathogen interactions, involve a molecular arms race. When plants mount a defense response against an unwanted viral invasion, the invaders eventually evolve mechanisms to thwart or sabotage the defense response, thus placing plants back under pressure to evolve immunity to the saboteurs. One component of the plant defense response that is at the forefront of this molecular arms race is autophagy (Han et al., 2015). Autophagy is a targeted degradation and recycling pathway in plants activated under nutritional deprivation, but its network of autophagosomes can also perform the targeted removal of unwanted cellular components. In several cases, viruses interfere with and suppress host autophagy because it is targeting viral factors for degradation and thus hindering infection (Kushwaha et al., 2019). In other cases, pathogens have evolved to hijack autophagy to degrade host defense components, such as the RNA silencing pathway, for their own benefit (Kushwaha et al., 2019). However, the Cotton leaf curl Multan virus (CLCuMuV) investigated by researchers in the lab of Yule Liu, induces autophagy in the host Nicotiana benthamiana that in turn targets a key viral protein, βC1, for degradation (Haxim et al., 2017). From an evolutionary perspective, such behaviour is perplexing as it is unclear who benefits (cui bono) from this induction of autophagy: the virus or the plant.
 To better understand the interaction between viruses and autophagy, Ismayil et al. (2020) sought to determine how CLCuMuV induces autophagy. CLCuMuV exists in association with the satellite DNA Cotton leaf curl Multan betasatellite (CLCuMuB), which encodes the single pathogenicity protein βC1. CLCuMuB βC1 suppresses RNA silencing, a common anti-viral defense mechanism in plants, and is therefore required for CLCuMuV infection to become symptomatic. Furthermore, βC1 is targeted by autophagy in CLCuMuV infected N. Benthamiana cells through its interaction with the autophagosome cargo-recruitment protein ATG8 (Haxim et al., 2017). It was, however, unknown which viral factors were inducing this autophagy. When Ismayil et al. expressed the βC1 protein in N. benthamiana in the absence of viral infection, these plants displayed a threefold increase in autophagosome formation, indicating that βC1 on its own induces autophagy. The authors then hypothesized that the βC1-ATG8 interaction was responsible for inducing autophagy, but they disproved this idea by observing that a mutated βC1 that does not bind ATG8 still induces autophagy.
To better understand the interaction between viruses and autophagy, Ismayil et al. (2020) sought to determine how CLCuMuV induces autophagy. CLCuMuV exists in association with the satellite DNA Cotton leaf curl Multan betasatellite (CLCuMuB), which encodes the single pathogenicity protein βC1. CLCuMuB βC1 suppresses RNA silencing, a common anti-viral defense mechanism in plants, and is therefore required for CLCuMuV infection to become symptomatic. Furthermore, βC1 is targeted by autophagy in CLCuMuV infected N. Benthamiana cells through its interaction with the autophagosome cargo-recruitment protein ATG8 (Haxim et al., 2017). It was, however, unknown which viral factors were inducing this autophagy. When Ismayil et al. expressed the βC1 protein in N. benthamiana in the absence of viral infection, these plants displayed a threefold increase in autophagosome formation, indicating that βC1 on its own induces autophagy. The authors then hypothesized that the βC1-ATG8 interaction was responsible for inducing autophagy, but they disproved this idea by observing that a mutated βC1 that does not bind ATG8 still induces autophagy.
The authors therefore expanded their search for other βC1 interactions that might induce autophagy. Using affinity purification followed by liquid chromatography-tandem mass spectrometry, they identified several cytosolic isoforms of glyceraldehyde-3-phosphate dehydrogenase (GAPC) as βC1 interacting proteins. This interaction is intriguing because, besides its role as a glycolytic enzyme, GAPC also acts as a signalling factor controlling autophagy via its interaction with autophagy-related protein ATG3 (Han et al., 2015). The βC1-GAPC interaction was further detailed in a series of co-immunopurification and bimolecular fluorescence complementation experiments that demonstrated that βC1 disrupted the interaction between GAPC and ATG3, which relieved the repressive effect of GAPC on autophagy. Together, these results provide a detailed mechanism of viral induction of autophagy in plants.
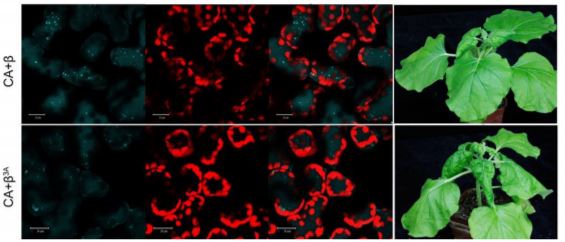
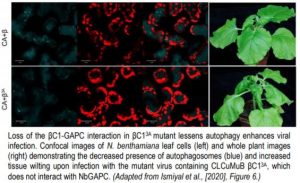
Plant infected with mutant versions of the virus, encoding a modified βC1 protein with impaired ability to interact with GAPC, displayed less autophagy, increased viral propagation and increased symptoms of infection (see Figure). This result is consistent with a previous finding that a mutant βC1 that avoided degradation by autophagy also increased CLCuMuV propagation in planta (Haxim et al., 2017). So why has the CLCuMuB βC1 protein evolved to initiate its own destruction thereby limiting its anti-defense-response activity? The authors hypothesize that by mediating self-autophagy, βC1 is moderating its virulence in an attempt to optimize host cell survival for better viral propagation and transmission. The verdict must therefore be that βC1 induces autophagy for its own benefit; otherwise such a mechanism would have been selected against. However, this ruling may yet be overturned as the situation is still evolving!
Brendan M O’Leary
ARC Centre of Excellence in Plant Energy Biology
University of Western Australia
ORCID: 0000-0002-8770-155X
References:
Han, S., Wang, Y., Zheng, X., Jia, Q., Zhao, J., Bai, F., Hong, Y., and Liu, Y. (2015). Cytoplastic Glyceraldehyde-3-Phosphate Dehydrogenases Interact with ATG3 to Negatively Regulate Autophagy and Immunity in Nicotiana benthamiana. Plant Cell 27: 1316-1331.
Haxim, Y., Ismayil, A., Jia, Q., Wang, Y., Zheng, X., Chen, T., Qian, L., Liu, N., Wang, Y., Han, S., Cheng, J., Qi, Y., Hong, Y., and Liu, Y. (2017). Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife 6:e23897.
Ismayil, A., Yang, M., Haxima, Y., Wang, Y., Li, J., Han, L., Wang, Y., Zheng, X., Wei, X., Nagalakshmi, U., Hong, Y., Hanley-Bowdoin, L., and Liu, Y. Cotton leaf curl Multan virus βC1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with Glyceraldehyde-3-Phosphate Dehydrogenases. Plant Cell https://doi.org/10.1105/tpc.19.00759.
Kushwaha, N.K., Hafren, A., and Hofius, D. (2019). Autophagy-virus interplay in plants: from antiviral recognition to proviral manipulation. Mol. Plant Pathol. 20: 1211-1216.