The AREB1-ADA2b-GCN5 Complex Regulates Gene Expression during Drought Stress
Abiotic stresses constitute a global threat to agricultural crop production and natural ecosystems. One of the most prominent abiotic stresses is drought, which dramatically alters plant physiology and morphology. Studies in model organisms have shed light into how plants respond to drought stress, including transcriptional reprogramming of genes responsive to the hormone abscisic acid (ABA) (Song et al., 2016), and epigenetic modifications, like histone acetylation (Kim et al., 2008). Exciting results by Li et al. (2018) further illuminate our understanding by mechanistically linking plant drought tolerance and resilience with ABA-responsive gene activation through differential histone modifications in the tree species Populus trichocarpa.
To gain an understanding of the global histone acetylation landscape in P. trichocarpa during drought stress, the authors performed ChIP-Seq of well-watered and drought-stressed plants using antibodies against acetylated histone H3 at lysine 9 (H3K9ac), which is associated with an open chromatin and is a known activation mark for gene expression. Using the ChIP-Seq results vis-a-vis parallel RNA-Seq analyses, they found that genomewide changes in both H3K9ac profiles and transcript levels were heavily associated with ABA signaling pathway-related genes. In these genes of interest, there was significant enrichment of the ABA-Responsive Element (ABRE) promoter motif that could be bound by the AREB1 (ABA-Responsive Element Binding 1)-type transcription factor (Fujita et al., 2005).
Because ABA-associated genes encoding transcription factors (TFs) have been shown to be effective for drought tolerance, the genes of interest were narrowed down to TF genes that met three important criteria: (1) differentially expressed during drought stress; (2) contained the ABRE motif; and (3) possessed changes in promoter H3K9ac levels. From this set of TF genes, Li et al. (2018) systematically narrowed them further to those with orthologs in other plant species that can enhance drought tolerance. Ultimately, three were selected as representative Populus TF genes for detailed functional characterization – PtrNAC006, –007 and –120. When these PtrNAC genes were overexpressed, transgenics lines had enhanced drought tolerance and resilience. The plants not only recovered after water deprivation but also resumed the same growth and development as their well-watered counterparts after recovery.
 But what is the mechanism of drought tolerance and resilience? For this important question, the authors examined changes in physiology and morphology. PtrNAC-overexpressing plants had increased water potential during drought stress. Additionally, their xylem vessels had reduced sizes and higher numbers compared to wild-type controls. Probing deeper into the molecular mechanisms linking ABA-responsive gene expression and drought tolerance, a protoplast system (Lin et al., 2014) was used to show that the Populus ortholog of AREB1 protein (PtrAREB1-2) can activate the transcription of the drought tolerance-associated PtrNAC006, –007 and –120 genes. ChIP and electrophoretic mobility shift assay (EMSA) confirmed that PtrAREB1-2 binds to the ABRE promoter motifs of these PtrNAC genes.
But what is the mechanism of drought tolerance and resilience? For this important question, the authors examined changes in physiology and morphology. PtrNAC-overexpressing plants had increased water potential during drought stress. Additionally, their xylem vessels had reduced sizes and higher numbers compared to wild-type controls. Probing deeper into the molecular mechanisms linking ABA-responsive gene expression and drought tolerance, a protoplast system (Lin et al., 2014) was used to show that the Populus ortholog of AREB1 protein (PtrAREB1-2) can activate the transcription of the drought tolerance-associated PtrNAC006, –007 and –120 genes. ChIP and electrophoretic mobility shift assay (EMSA) confirmed that PtrAREB1-2 binds to the ABRE promoter motifs of these PtrNAC genes.
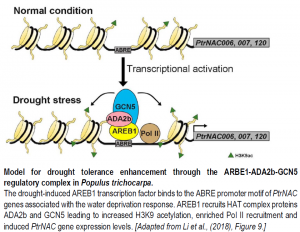
Because these PtrNAC genes also had differential H3K9ac levels in their promoter regions, it was important to test if the PtrAREB1-2 transcriptional activator can associate with a histone acetyltransferase (HAT) complex. Remarkably, using in vitro and in vivo protein interaction assays, PtrAREB1-2 was found to interact with the HAT complex catalytic subunit PtrGCN5 and the adaptor protein PtrADA2b. The P. trichocarpa AREB1-ADA2b-GCN5 regulatory complex dramatically induced transcription of the three PtrNAC genes by increasing H3K9ac levels and RNA polymerase II recruitment. Functional characterization of the ternary complex proteins by RNAi silencing and CRISPR editing provided convincing evidence that PtrAREB1, PtrADA2b, and PtrGCN5 are required for drought tolerance.
Overall, this study by Li et al. (2018) is a comprehensive investigation into the molecular mechanisms leading to plant responses to drought stress. The authors successfully connected drought tolerance responses (like higher water potential) with ABRE-directed gene expression and H3K9ac enrichment. In the future, it would be interesting to characterize the downstream target genes of the PtrNAC transcription factors leading to enhanced drought tolerance, and also upstream cellular signals leading to the formation of the AREB1-ADA2b-GCN5 ternary complex.
REFERENCES
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 17: 3470-3488.
Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, Toyoda T, Kimura H, Shinozaki K, Seki M (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49: 1580-1588. doi: 10.1093/pcp/pcn133.
Li S, Lin YCJ, Wang P, Zhang B, Li M, Chen S, Shi R, Tunlaya-Anukit S, Liu X, Wang Z, Dai X, Yu J, Zhou C, Liu B, Wang JP, Chiang VL, Li W (2018). Histone acetylation cooperating with AREB1 transcription factor regulates drought response and tolerance in Populus trichocarpa. Plant Cell. https://doi.org/10.1105/tpc.18.00437
Lin YC, Li W, Chen H, Li Q, Sun YH, Shi R, Lin CY, Wang JP, Chen HC, Chuang L, Qu GZ, Sederoff RR, Chiang VL (2014). A simple improved-throughput xylem protoplast system for studying wood formation. Nat. Protoc. 9: 2194-2205.
Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR. (2016). A transcription factor hierarchy defines an environmental stress response network. Science. 354 (6312). pii: aag1550.



