TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis (PNAS)
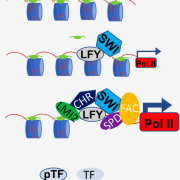
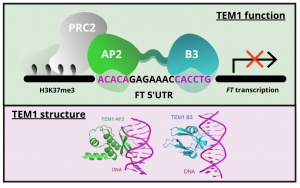 Gene expression is primarily controlled by transcriptional regulatory proteins able to recognize and bind short DNA sequences in downstream targets. Understanding how DNA-binding proteins achieve high precision and switch on/off the transcription of selected genes at a specific developmental stage or in response to a specific environmental stimulus has been one of the major goals of research in the field of molecular biology. In this paper, Hu and colleagues investigated the structure and function of the floral repressor TEMPRANILLO1 (TEM1), characterized by an N-terminal AP2 and central B3 domains. In Arabidopsis, TEM1 negatively regulates flowering time under non inductive conditions by directly repressing the florigen FLOWERING LOCUS T (FT). The authors first determined the crystal structure of the AP2 and B3 DNA binding domains and identified residues involved in the recognition of cis-acting elements present in FT 5’UTR. They also analyzed the affinity between different versions of TEM1 and a DNA probe containing both AP2 and B3 elements. In vitro binding assays revealed that the combination of the two protein domains increased affinity to the target sequence, while mutations in key residues or deletions disrupted the DNA-protein interaction. In planta, the expression of mutated or deleted forms of TEM1 failed to rescue the early flowering phenotype of tem mutants, suggesting that both domains are required to regulate the florigen. TEM1 also interacts with the catalytic subunit of the Polycomb Repressor Complex 2 (PRC2), responsible for the deposition of repressive marks at target loci. Thus, TEM1 binds to specific sequences of FT and recruits chromatin remodeling factors to trigger its epigenetic silencing. (Summary and image adaptation by Michela Osnato @michela_osnato) PNAS 10.1073/pnas.2103895118
Gene expression is primarily controlled by transcriptional regulatory proteins able to recognize and bind short DNA sequences in downstream targets. Understanding how DNA-binding proteins achieve high precision and switch on/off the transcription of selected genes at a specific developmental stage or in response to a specific environmental stimulus has been one of the major goals of research in the field of molecular biology. In this paper, Hu and colleagues investigated the structure and function of the floral repressor TEMPRANILLO1 (TEM1), characterized by an N-terminal AP2 and central B3 domains. In Arabidopsis, TEM1 negatively regulates flowering time under non inductive conditions by directly repressing the florigen FLOWERING LOCUS T (FT). The authors first determined the crystal structure of the AP2 and B3 DNA binding domains and identified residues involved in the recognition of cis-acting elements present in FT 5’UTR. They also analyzed the affinity between different versions of TEM1 and a DNA probe containing both AP2 and B3 elements. In vitro binding assays revealed that the combination of the two protein domains increased affinity to the target sequence, while mutations in key residues or deletions disrupted the DNA-protein interaction. In planta, the expression of mutated or deleted forms of TEM1 failed to rescue the early flowering phenotype of tem mutants, suggesting that both domains are required to regulate the florigen. TEM1 also interacts with the catalytic subunit of the Polycomb Repressor Complex 2 (PRC2), responsible for the deposition of repressive marks at target loci. Thus, TEM1 binds to specific sequences of FT and recruits chromatin remodeling factors to trigger its epigenetic silencing. (Summary and image adaptation by Michela Osnato @michela_osnato) PNAS 10.1073/pnas.2103895118