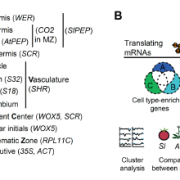
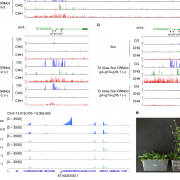
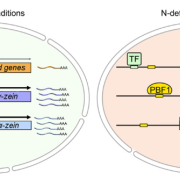
Systemic RNA Silencing in Tobacco
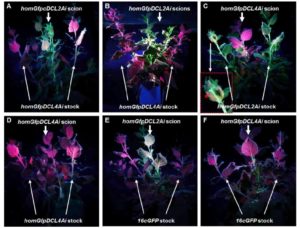 gene RNA silencing is a cellular gene regulatory mechanism that is conserved across fungal, plant, and animal kingdoms. Through sequence-specific targeting, RNA silencing can degrade mRNA for posttranscriptional gene silencing (PTGS) or modify related DNA for transcriptional gene silencing. RNA silencing can spread from cell to cell and over long distances in animals and plants. However, the genetic requirements and signals involved in plant mobile gene silencing are poorly understood. In plants, primary silencing is triggered by double-stranded RNA (dsRNA) molecules, which are processed into 21 to 24 nucleotide (nt) small-interfering (si) RNA duplexes, known as primary siRNAs, by DICER-LIKE (DCL) RNase III-like enzymes. Chen et al. (10.1104/pp.17.01828) have identified a DICER-LIKE2 (DCL2)-dependent mechanism for systemic spread of PTGS in Nicotiana benthamiana. As in Arabidopsis, DCL1 is involved in microRNA production, whereas DCL4, DCL2, and DCL3 are required to produce 21, 22, and 24 nt siRNAs, respectively. Inhibition of DCL2 expression reduced the spread of gene silencing, while suppression of DCL3 or DCL4 expression enhanced systemic PTGS. The authors further show that the 21 or 24 nt local siRNAs produced by DCL4 or DCL3 were not involved in long-distance gene silencing. Grafting experiments demonstrated that DCL2 was required in the scion to respond to the signal, but not in the rootstock to produce/send the signal. These results suggest a coordinated DCL genetic pathway in N. benthamiana in which DCL2 plays an essential role in systemic PTGS. DCL4 and DCL3, on the hand, reduce systemic PTGS.
gene RNA silencing is a cellular gene regulatory mechanism that is conserved across fungal, plant, and animal kingdoms. Through sequence-specific targeting, RNA silencing can degrade mRNA for posttranscriptional gene silencing (PTGS) or modify related DNA for transcriptional gene silencing. RNA silencing can spread from cell to cell and over long distances in animals and plants. However, the genetic requirements and signals involved in plant mobile gene silencing are poorly understood. In plants, primary silencing is triggered by double-stranded RNA (dsRNA) molecules, which are processed into 21 to 24 nucleotide (nt) small-interfering (si) RNA duplexes, known as primary siRNAs, by DICER-LIKE (DCL) RNase III-like enzymes. Chen et al. (10.1104/pp.17.01828) have identified a DICER-LIKE2 (DCL2)-dependent mechanism for systemic spread of PTGS in Nicotiana benthamiana. As in Arabidopsis, DCL1 is involved in microRNA production, whereas DCL4, DCL2, and DCL3 are required to produce 21, 22, and 24 nt siRNAs, respectively. Inhibition of DCL2 expression reduced the spread of gene silencing, while suppression of DCL3 or DCL4 expression enhanced systemic PTGS. The authors further show that the 21 or 24 nt local siRNAs produced by DCL4 or DCL3 were not involved in long-distance gene silencing. Grafting experiments demonstrated that DCL2 was required in the scion to respond to the signal, but not in the rootstock to produce/send the signal. These results suggest a coordinated DCL genetic pathway in N. benthamiana in which DCL2 plays an essential role in systemic PTGS. DCL4 and DCL3, on the hand, reduce systemic PTGS.