Sweeter than SWEET: A Single-Cell Leaf Vasculature Transcriptome Atlas
Kim et al. provide insight into leaf vascular cell types and new resources to build hypotheses and strategies regarding transport of ions, metabolites, and signals.
Plant Cell http://bit.ly/2MKOR8M
Ji-yun Kim and Wolf Frommer
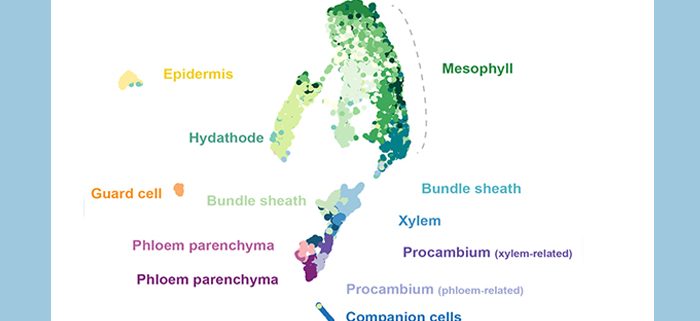
Background: Transport processes are critical for all multicellular organisms. In plants, two vascular tissues, phloem and xylem, are mainly responsible for fulfilling this task. The leaf vascular cells are embedded deep inside the leaf and were mostly identified microscopically. Cell type specific markers are rare for most vascular cell types. Especially, little is known about the phloem parenchyma, a cell type of the phloem involved in sucrose transport. To gain more profound knowledge of the transcript profiles of the leaf vascular cells, we optimized protocols to enrich vascular protoplasts (plant cells without cell walls) and performed single-cell RNA sequencing analysis.
Questions: What is the mRNA profile of different vascular cell types? Can we identify the transcriptome of the phloem parenchyma? What types of transporters are represented in different cell types? Which metabolic pathways are enriched in vascular cell types?
Findings: Our single-cell transcriptome atlas contains all cell types in the Arabidopsis leaf vasculature and non-vascular cell types such as bundle sheath, epidermis, guard cells, and mesophyll cells. We were able to identify distinct mRNA signatures which reflect vascular cell type identity. We found that the phloem parenchyma cells are enriched for various transporters, including sugar transporters and amino acid transporters. In a parallel article in this issue, we show that maize homologs of these sugar and amino acid transporters are enriched in the abaxial bundle sheath cells. These observations suggest that maize and Arabidopsis use different routes for phloem loading of sucrose and amino acids. In addition, we found unexpected roles of PP in hormone biosynthesis and defense-related responses.
Next steps: As we now have the resources at hand, our next step will be to explore the vascular cell types at the molecular level to dissect their function. Our goal is to identify specific components, for instance, transcription factors or transport proteins and contribute to the solution of the long-standing riddles: How do plants distribute nutrients across the entire plant body and how are the logistics coordinated?
Ji-Yun Kim, Efthymia Symeonidi, Tin Yau Pang, Tom Denyer, Diana Weidauer, Margaret Bezrutczyk, Manuel Miras, Nora Zöllner, Thomas Hartwig, Michael M. Wudick, Martin Lercher, Li-Qing Chen, Marja C.P Timmermans & Wolf B. Frommer. (2021). Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell. DOI: 10.1093/plcell/koaa060