Starting Off Right: N-terminal Acetylation Stabilizes an Immune-activating Protein
Charles Copeland1
1Department of Plant Microbe Interactions, Max Planck Institute for Plant Breeding Research, 50829 Cologne, Germany
Plants rely on a complex innate immune system to recognize and respond to potential pathogens in their environment (Jones and Dangl, 2006). Activation of immune responses generally prevents infection; however, immune response activation is also detrimental for plant growth, and must be tightly regulated to avoid unnecessary negative effects in the absence of a pathogen threat. Immune-related proteins can be regulated at both transcriptional and post-transcriptional levels. However, much remains to be learned about the specificity of this regulation, and how different mechanisms of regulation combine to fine-tune the activity of these proteins.
 In this issue of Plant Physiology, Li et al (2020) explore how the stability of the Arabidopsis thaliana immune-related protein SIGMA FACTOR-BINDING PROTEIN 1 (SIB1) is affected by a post-translational modification, N-terminal acetylation. Traditionally, an N-terminal acetyl group is thought to be a destabilizing feature, marking proteins for degradation through the N-end rule pathway (Dissmeyer et al., 2018). In this pathway, certain N-terminal amino acid residues, in some cases modified by acetylation, are recognized by E3 ligase enzymes. E3 ligases add a ubiquitin peptide onto a nearby lysine residue, triggering degradation by the proteasome complex. However, here the authors found that N-terminal acetylation of SIB1 actually promotes its stability, indicating that the regulation of protein turnover through the N-end rule pathway is more complex than previously thought.
In this issue of Plant Physiology, Li et al (2020) explore how the stability of the Arabidopsis thaliana immune-related protein SIGMA FACTOR-BINDING PROTEIN 1 (SIB1) is affected by a post-translational modification, N-terminal acetylation. Traditionally, an N-terminal acetyl group is thought to be a destabilizing feature, marking proteins for degradation through the N-end rule pathway (Dissmeyer et al., 2018). In this pathway, certain N-terminal amino acid residues, in some cases modified by acetylation, are recognized by E3 ligase enzymes. E3 ligases add a ubiquitin peptide onto a nearby lysine residue, triggering degradation by the proteasome complex. However, here the authors found that N-terminal acetylation of SIB1 actually promotes its stability, indicating that the regulation of protein turnover through the N-end rule pathway is more complex than previously thought.
SIB1 acts as a transcriptional co-regulator with both nuclear and chloroplastic localization. In the nucleus, it enhances the DNA binding activity of the transcription factor WRKY33 to induce expression of immune-related genes (Lai et al., 2011). In addition, the nuclear and chloroplastic forms of SIB1 differentially affect the expression photosynthesis-related genes, leading to an imbalance in chloroplast function which can promote runaway cell death (Lv et al., 2019). Thus, SIB1 levels must be tightly controlled.
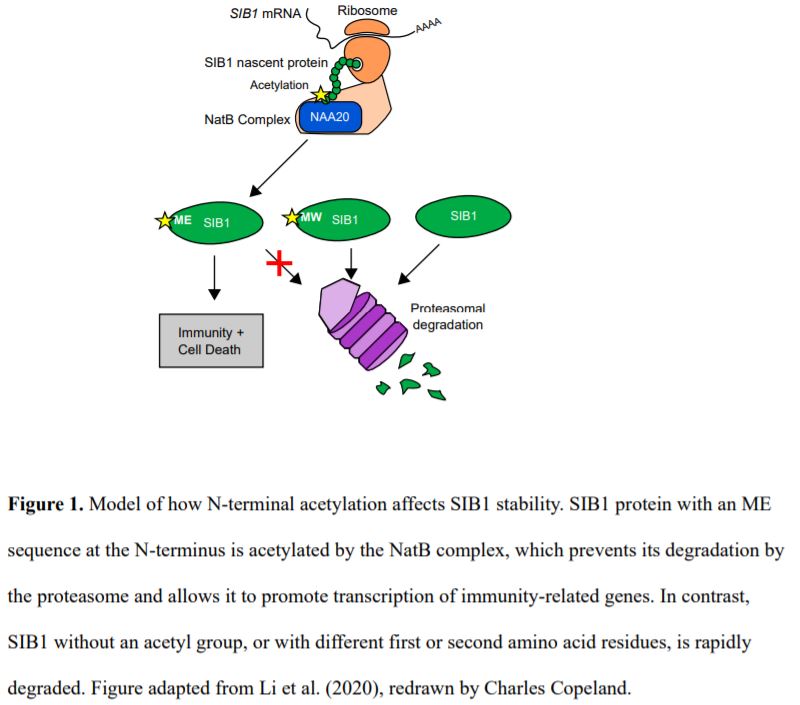
In agreement with previous reports, Li et al. (2020) found that SIB1 expression is extremely low under normal growth conditions, resulting in an almost negligible protein abundance. However, after treatment with the defense hormone salicylic acid (SA), SIB1 is induced, and protein levels rise rapidly. They also found that the nuclear form of SIB1 protein was much larger than expected, indicating possible post-translational modifications. Mass spectrometry showed SIB1 is always acetylated at the initial methionine (M) residue. The presence of a glutamate (E) residue at the second position suggested that the acetylation is performed by the NatB complex. Indeed, the naa20 mutant, which is defective in the catalytic subunit of NatB, contains a mix of acetylated and non-acetylated SIB1, confirming that NatB is partially responsible for the acetylation (Fig. 1). The Arabidopsis genome contains a number of other Nat complexes, and it remains to be seen which of these is responsible for the remaining N-terminal acetylation of SIB1.
Silencing of NAA20 in Nicotiana benthamiana reduced the N-terminal acetylation of ectopically expressed SIB1, accompanied by a much lower abundance of SIB1 protein but not SIB1 transcripts. The reduced protein abundance was surprising, since N-terminal acetylation is usually thought to mark proteins for degradation. To further analyze the effect of N-terminal acetylation on SIB1 stability, the authors created SIB1 constructs encoding for the protein with different amino acid residues at the second position, designed to abolish the acetylation or trigger recognition by different E3 ligases. All of the altered SIB1 proteins were much less stable. Thus, both the acetylation and the ME sequence at the N-terminus of SIB1 are required to save it from degradation.
The authors next examined the importance of SIB1 N-terminal acetylation on its biological function, which is most easily observed in the lesion simulating disease1 (lsd1) mutant background. Mature lsd1 plants show upregulation of defense marker genes, and lesions caused by runaway cell death. However, these phenotypes are suppressed in the lds1 sib1 double mutant. Expression of the SIB1 constructs with altered N-termini could not restore the marker gene expression or cell death. Since the substituted proteins do not have altered nuclear localization or interaction with WRKY33, this indicates that SIB1 protein cannot accumulate to functional levels without N-terminal acetylation (Fig. 1).
Although acetylation of the N-terminus generally acts as a degradation signal, it remains difficult to predict which proteins are actually targeted by the N-end rule pathway (Dissmeyer et al., 2018). Acetylated proteins may escape degradation if the acetyl group is not accessible to the E3 ligase, or if there is no suitable lysine residue for ubiquitin conjugation. However, this does not seem to be the case for SIB1, making it even more intriguing that N-terminal acetylated SIB1 is stabilized (Li et al., 2020). Another Arabidopsis immune-related protein, SNC1, is also stabilized by specific N-terminal acetylation (Xu et al., 2015). In this case, the stabilized protein begins with an initial M followed by an aspartic acid (D) residue, similar to the ME sequence of SIB1, and is likewise acetylated by NatB. Further study is needed to determine how an acetylated ME or MD N-terminal sequence could affect a protein’s interactions with E3 ligases or other components of proteasomal degradation pathway.
Modulation of protein stability by the N-end rule pathway has been shown to mediate plant responses against specific stresses. For example, hypoxia-responsive transcription factors, normally targets of the N-end rule pathway, become modified at the N-terminus under low oxygen conditions, preventing their recognition and degradation (Gibbs et al., 2011). The biological significance of stabilizing SIB1 through N-terminal acetylation is less clear. The activity of immunity-promoting proteins should certainly be restricted unless a pathogen threat is present. However, SIB1 is already strongly regulated by SA at the transcriptional level, whereas NatB activity is not obviously affected (Li et al., 2020). Interestingly, proteasome activity increases during the plant immune response, resulting in more protein degradation (Üstün et al., 2016). Blocking the proteasome pathway results in enhanced disease susceptibility, indicating that protein degradation contributes to disease resistance. At the same time, induced immunity-activating proteins must be able to accumulate, and it is possible that such accumulation could be achieved through specific N-terminal acetylation. Overall, the findings of Li et al. (2020) show that N-terminal acetylation does not simply serve as a mark for protein degradation, but can actually stabilize proteins such as SIB1. It would be interesting to examine the N-terminal acetylation state of a larger set of immune-related proteins, as well as its effect on their stability, to determine whether this phenomenon is more widespread.
Literature Cited
Dissmeyer N, Rivas S, Graciet E (2018) Life and death of proteins after protease cleavage: protein degradation by the N-end rule pathway. New Phytol 218: 929–935
Gibbs DJ, Lee SC, Md Isa N, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, et al (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329
Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z (2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841
Lv R, Li Z, Li M, Dogra V, Lv S, Liu R, Lee KP, Kim C (2019) Uncoupled expression of nuclear and plastid photosynthesis-associated genes contributes to cell death in a lesion mimic mutant. Plant Cell 31: 210–230
Üstün S, Sheikh A, Gimenez-Ibanez S, Jones A, Ntoukakis V, Börnke F (2016) The Proteasome Acts as a Hub for Plant Immunity and Is Targeted by Pseudomonas Type III Effectors. Plant Physiol 172: 1941–1958
Xu F, Huang Y, Li L, Gannon P, Linster E, Huber M, Kapos P, Bienvenut W, Polevoda B, Meinnel T, et al (2015) Two N-Terminal Acetyltransferases Antagonistically Regulate the Stability of a Nod-Like Receptor in Arabidopsis. Plant Cell 27: 1–17