Identifying and annotating mRNA transcripts from non-genic regions of the genome
Panda et al. use third-generation sequencing to examine the transcriptome of genomic regions that do not encode genes. Plant Cell https://doi.org/10.1105/tpc.20.00115
By Kaushik Panda – Donald Danforth Plant Science Center, and R Keith Slotkin – Donald Danforth Plant Science Center and University of Missouri
Background: In spite of making up only a small percentage of plant or animal genomes, genes are the focal point of most molecular biology research. The rest of the genome is often excluded from analysis simply due to lack of adequate resources. For example, there is no annotation of the RNA transcripts generated from non-genic regions.
Questions: In addition to genes, which regions of the genome can produce functional mRNAs? How will the identification and annotation of such mRNAs improve the analysis of these regions using existing tools?
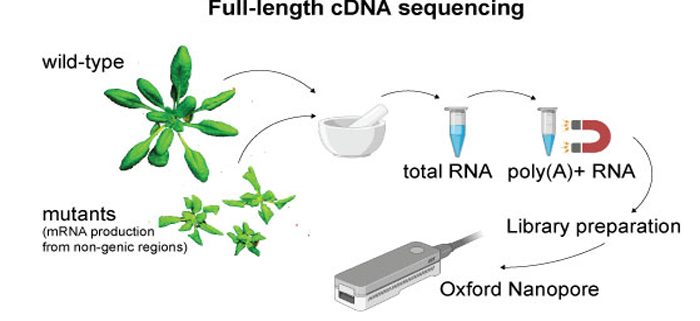
Findings: In this study, we have used third-generation sequencing of full-length mRNAs on plants that transcribe these non-genic regions of the genome. The product is an annotation of mRNA transcripts generated from the previously overlooked territories of the genome. We use this new annotation to demonstrate how other experiments can now investigate the entire genome, rather than just the genes.
Next steps: We used mutants to uncover otherwise repressed non-genic mRNAs in two well-studied plant genomes. A wider breadth of mRNAs can be captured, including in more diverse species, by investigating different tissues or combining stress treatments with mutants.
Kaushik Panda and R. Keith Slotkin (2020) Long-read cDNA Sequencing Enables a ‘Gene-Like’ Transcript Annotation of Transposable Elements. https://doi.org/10.1105/tpc.20.00115