Spotlight: Super-pangenomes for improved breeding
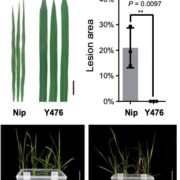
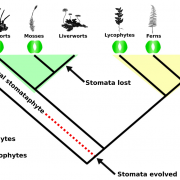
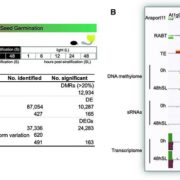
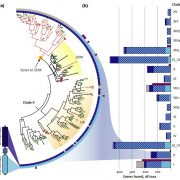
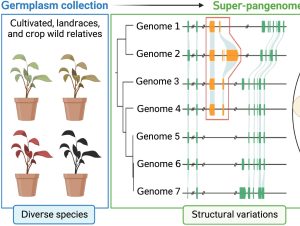 Sometimes more really is better, and I think it’s safe to say that when it comes to genomic information, more is better. Here, Raza et al. highlight the great value of super-pangenomes. A pan-genome is defined as the entire set of genes within a species, created by combining sequences of many individuals. The super-pangenome as defined here expands this concept to incorporate genomes of close relatives, up to the genus level. This idea is particularly useful as a way to incorporate traits from crop wild relatives into breeding programs. Crop wild relatives often are sources of genes the provide environmental robustness that has been lost through selection for yields, but introgressions of these valuable traits can be difficult because of structural variations that have arisen over time. The authors cite examples of super-pangenome assemblies that have been developed for rice, soybean, and tomato, and how these resources have allowed the identification of several novel genes. Several strategies for breeding using information from super-pangenomes is also discussed. (Summary by Mary Williams @PlantTeaching) Mol. Plant 10.1016/j.molp.2023.08.005
Sometimes more really is better, and I think it’s safe to say that when it comes to genomic information, more is better. Here, Raza et al. highlight the great value of super-pangenomes. A pan-genome is defined as the entire set of genes within a species, created by combining sequences of many individuals. The super-pangenome as defined here expands this concept to incorporate genomes of close relatives, up to the genus level. This idea is particularly useful as a way to incorporate traits from crop wild relatives into breeding programs. Crop wild relatives often are sources of genes the provide environmental robustness that has been lost through selection for yields, but introgressions of these valuable traits can be difficult because of structural variations that have arisen over time. The authors cite examples of super-pangenome assemblies that have been developed for rice, soybean, and tomato, and how these resources have allowed the identification of several novel genes. Several strategies for breeding using information from super-pangenomes is also discussed. (Summary by Mary Williams @PlantTeaching) Mol. Plant 10.1016/j.molp.2023.08.005