Some Things Never Change: Conserved MYC-family bHLH Transcription Factors Mediate dinor-OPDA Signaling in Liverworts
The lipidic phytohormone jasmonyl-isoleucine (JA-Ile) is a key mediator of stress-versus-growth signaling in vascular plants. Upon it’s accumulation, JA-Ile is detected by the F box receptor protein COI1 (CORONATINE INSENSITIVE1), which in turn leads to the ubiquitination and proteasomal degradation of JAZ (JASMONATE ZIM-DOMAIN) repressor proteins that would otherwise bind to and inhibit MYC-family bHLH transcription factors. This hormone-triggered de-repression of the jasmonate (JA) pathway allows for the induction of JA-related gene expression, physiological adjustments, and the accumulation of specialized metabolites like glucosinolates and terpenoids (Wasternack and Hause 2013).
Studies exploring the evolutionary history and functional significance of JA signaling in early divergent land plants (bryophytes) have recently uncovered a conserved COI1-JAZ module centered on the perception of the modified JA and OPDA (12-oxo phytodienoic acid) precursor dinor-OPDA (dn-OPDA) rather than JA-Ile (Monte et al., 2018; Monte et al., 2019). In this issue, Peñuelas et al. (2019) build on this knowledge and determine the degree to which JA/dn-OPDA signaling in bryophytes relies on the conserved action of MYC-family bHLH transcription factors. Phylogenetic analyses identified MYC homologs with appropriate domain architecture in certain charophytic (freshwater) algal lineages and across land plants, indicating that MYC transcription factors predate the colonization of land. In the model liverwort Marchantia polymorpha, two ubiquitously expressed MpMYC homologs were discovered on male (MpMYCY) or female (MpMYCX) sex chromosomes. Both MpMYC homologs interact strongly with the MpJAZ repressor through conserved interaction domains, similar to angiosperm MYC and JAZ homologs. Moreover, Marchantia MpMYC homologs interact with Arabidopsis AtJAZ proteins, suggesting a strong evolutionary pressure to maintain MYC-JAZ interactions throughout the evolution of plants.
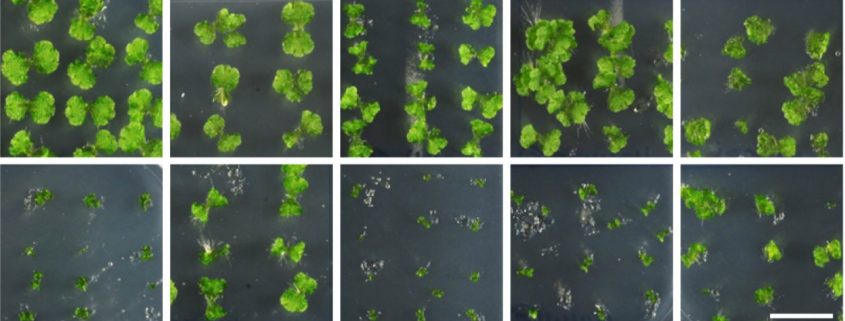
 Phenotypic analysis of overexpression and knockout lines revealed a functionally conserved role for MpMYC transcription factors in mediating dn-OPDA signaling in Marchantia. Compared to wild-type liverworts, Mpmyc knockouts were insensitive to the growth suppressing effects of exogenous OPDA, whereas MpMYC overexpressors displayed heightened sensitivity (see Figure). Microarray-based transcriptomic profiling of wild type versus Mpmyc liverworts that were wounded or treated with exogenous OPDA further defined a set of MYC-dependent genes whose promoters were enriched for conserved G-box motifs associated with MYC-binding. This set of MpMYC-dependent loci included terpenoid biosynthesis genes, whose relevance to OPDA-mediated anti-herbivory responses was supported by an enhanced susceptibility to generalist insect larvae in Mpmyc mutants that could not accumulate sesquiterpene terpenoids (Peñuelas et al., 2019).
Phenotypic analysis of overexpression and knockout lines revealed a functionally conserved role for MpMYC transcription factors in mediating dn-OPDA signaling in Marchantia. Compared to wild-type liverworts, Mpmyc knockouts were insensitive to the growth suppressing effects of exogenous OPDA, whereas MpMYC overexpressors displayed heightened sensitivity (see Figure). Microarray-based transcriptomic profiling of wild type versus Mpmyc liverworts that were wounded or treated with exogenous OPDA further defined a set of MYC-dependent genes whose promoters were enriched for conserved G-box motifs associated with MYC-binding. This set of MpMYC-dependent loci included terpenoid biosynthesis genes, whose relevance to OPDA-mediated anti-herbivory responses was supported by an enhanced susceptibility to generalist insect larvae in Mpmyc mutants that could not accumulate sesquiterpene terpenoids (Peñuelas et al., 2019).
Reciprocal cross-species complementation assays failed to rescue the JA/OPDA insensitivity phenotypes of Arabidopsis or Marchantia myc mutants, suggesting partial divergence between MYC signaling mechanisms in distantly related plant lineages. Upon further analysis, the authors discovered that MYC homologs could not interact with MED25 mediator subunits across species, rendering each homolog inactive when expressed out of context (Peñuelas et al., 2019). Such co-evolutionary constraints may be indicative of parallel evolutionary pressures required to fine-tune stress-versus-growth responses as plants diversified on land. In any case, the combined work of Peñuelas and colleagues now solidifies MYC transcription factors as broadly conserved master regulators of the JA/dn-OPDA pathway in land plants.
REFERENCES
Monte I, Ishida S, Zamarreno AM, Hamberg, Franco-Zorrilla JM, Garcia-Casado G, Gouhier-Darimont C, Reymond P, Takahashi K, Garcia-Mino JM, Nishihama R, Kohchi T and Solano R (2018). Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Bio. 14: 480-488.
Monte I, Franco-Zorrilla JM, Garcia-Casado G, Zamarreno A, Garcia-Mina JM, Nishihama R, Kohchi T and Solano R (2019). A single JAZ repressor controls the jasmonate pathway in Marchantia polymorpha. Mol. Plant 12: 185-198.
Peñuelas M, Monte I, Schweizer F, Vallat A, Reymond P, Garcia-Casado G, Franco-Zorilla JM and Solano R (2019). Functional conservation of jasmonate-related MYC transcription factors in Marchantia polymorpha. Plant Cell Published August 2019 DOI: https://doi.org/10.1105/tpc.19.00974.
Wasternack C and Hause B (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111: 1021-1058.
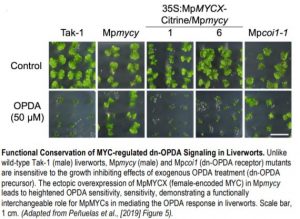
Figure legend:
Functional Conservation of MYC-regulated dn-OPDA Signaling in Liverworts. Unlike wild-type Tak-1 (male) liverworts, Mpmycy (male) and Mpcoi1 (dn-OPDA receptor) mutants are insensitive to the growth inhibiting effects of exogenous OPDA treatment (dn-OPDA precursor). The ectopic overexpression of MpMYCX (female-encoded MYC) in Mpmycy leads to heightened OPDA sensitivity, demonstrating a functionally interchangeable role for MpMYCs in mediating the OPDA response in liverworts. Scale bar, 1 cm. (Adapted from Peñuelas et al., [2019] Figure 5).