Slice and Dice: DCL2 Mediates the Production of 22-nt siRNAs That Influence Trait Variation in Soybean
In plants, small interfering RNAs (siRNAs) serve as key regulators of gene expression. While 24-nucleotide (nt) siRNAs are produced by DCL3 and mediate transcriptional silencing of transposons and pericentromeric chromatin through RdDM (Borges and Martienssen, 2015), 22-nt siRNAs are processed by DCL2 and participate in transgene silencing and viral defense (Wang et al., 2018; Parent et al., 2015). However, since 22-nt siRNAs are relatively rare in wild-type Arabidopsis (Henderson et al., 2006), and the loss of DCL2 doesn’t produce obvious development defects (Henderson et al., 2006; Wang et al., 2018), their role is still largely unknown. In contrast, crops such as soybean (Glycine max) exhibit abundant 22-nt siRNAs and are ideal systems to study their biogenesis, targeting and functions in genome activity.
In a new study, Jinbu Jia and co-authors combine genome editing, sRNA-seq and transcriptomics to investigate the functions of DCL2-dependent 22-nt siRNAs in soybean (Jia et al., 2020). The authors used CRISPR-Cas9 genome editing to simultaneously generate loss-of-function mutants of the two GmDLC2 genes in soybean, and performed sRNA-seq on a selection of tissues. The analysis revealed a substantial decrease in 22-nt siRNAs abundance throughout the genome in Gmdcl2, but also a drastic reduction of secondary 21-nt siRNAs derived from the 22-nt siRNAs. By investigating the distribution of these DCL2-dependent siRNAs in wild-type soybean, the authors identify that a large majority of 22-nt siRNAs are overlapping with transposable elements (TEs), suggesting a possible role in silencing. In the Gmdcl2 mutant, the production of such TE-associated 22-nt is drastically reduced, but does not correlate with local reduction in DNA methylation, nor accumulation of TE transcripts. Next, the authors examined the structural features of GmDCL2 substrates. The transcriptomic approach reveals that DCL2 preferentially process PolII-transcribed long inverted repeats to generate 22-nt siRNAs.
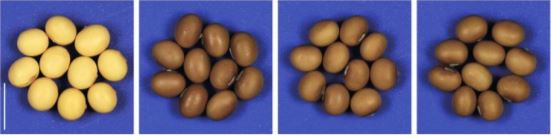
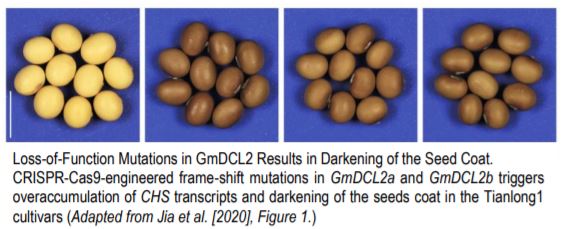 The most dramatic developmental phenotype resulting from GmDCL2 loss-of-function mutants is a darkening of the seed coat, from yellow in wild-type soybean to dark brown in Gmdlc2 (Figure). Seed color in soybean is dictated by the activity of Chalcone Synthase (CHS)-encoding genes that enable flavonoid biosynthesis and results in dark brown seed coat. The yellow seed coat phenotype is associated to the abundant production of siRNAs from a particular CHS gene cluster, which targets and silence other CHS genes in trans. With a higher genomic resolution provided by a de novo assembly of the CHS cluster, Jinbu Jia and colleagues propose that the CHS cluster generating siRNAS can form a long inverted repeat transcript, possibly processed by DCL2. This hypothesis is further supported by the accumulation of DCL2-dependent 22-nt siRNAs at this CHS cluster, which disappears in Gmdcl2. In absence of CHS trans-targeted siRNAs, other CHS genes accumulate transcripts at higher level compared to wild-type, and flavonoid accumulation results in dark seed coat.
The most dramatic developmental phenotype resulting from GmDCL2 loss-of-function mutants is a darkening of the seed coat, from yellow in wild-type soybean to dark brown in Gmdlc2 (Figure). Seed color in soybean is dictated by the activity of Chalcone Synthase (CHS)-encoding genes that enable flavonoid biosynthesis and results in dark brown seed coat. The yellow seed coat phenotype is associated to the abundant production of siRNAs from a particular CHS gene cluster, which targets and silence other CHS genes in trans. With a higher genomic resolution provided by a de novo assembly of the CHS cluster, Jinbu Jia and colleagues propose that the CHS cluster generating siRNAS can form a long inverted repeat transcript, possibly processed by DCL2. This hypothesis is further supported by the accumulation of DCL2-dependent 22-nt siRNAs at this CHS cluster, which disappears in Gmdcl2. In absence of CHS trans-targeted siRNAs, other CHS genes accumulate transcripts at higher level compared to wild-type, and flavonoid accumulation results in dark seed coat.
Further from facilitating trait diversification, DCL2-dependent 22-nt siRNAs are also involved in translational repression and stress adaptation (Wu et al., 2020). The high abundance of 22-nt siRNAs in crops suggests more unexplored functions in regulating genome activity in plants.
Matthias Benoit
Howard Hughes Medical Institute
Cold Spring Harbor Laboratory
ORCID: 0000-0002-3958-3173
REFERENCES
Borges, F. and Martienssen, R.A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16: 727–741.
Henderson, I.R., Zhang, X., Lu, C., Johnson, L., Meyers, B.C., Green, P.J., and Jacobsen, S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38: 721–725.
Parent, J.S., Jauvion, V., Bouché, N., Béclin, C., Hachet, M., Zytnicki, M., and Vaucheret, H. (2015). Post-transcriptional gene silencing triggered by sense transgenes involves uncapped antisense RNA and differs from silencing intentionally triggered by antisense transgenes. Nucleic Acids Res. 43: 8464–8475.
Wang, Z., Hardcastle, T.J., Pastor, A.C., Yip, W.H., Tang, S., and Baulcombe, D.C. (2018). A novel DCL2-dependent miRNA pathway in tomato affects susceptibility to RNA viruses. Genes Dev. 32: 1155–1160.
Wu, H. et al. (2020). Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature 581: 89–93.