Single seeds exhibit transcriptional heterogeneity during secondary dormancy induction (Plant Physiol)
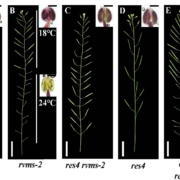
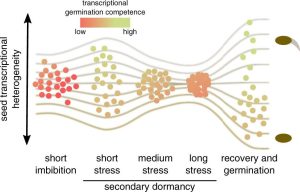 Secondary dormancy (SD) of seeds is a natural strategy that allows survival of the plant in environments with unfavourable climatic conditions. However, in Arabidopsis, a variance of dormancy depth in between identical seeds can be explained by population-based threshold models. This single-seed variability relies on the differences in oxygen consumption, hormone levels and sensitivity, and endo-β-mannanase enzymatic activity. In particular, the single-seed response to SD has been less studied than the response to primary dormancy. In this work, Krzyszton et al., describe a protocol for single-seed transcriptomic analysis to show differences between genetically identical seeds that are subjected to the same stress. The authors subjected Arabidopsis thaliana seeds to high temperature and darkness to induce SD. They observed that the treatment generates the same transcriptomic drop during the stress in all the seedlings. However, transcriptomic differences in the recovery step were observed between the germinating seeds and the ones that remained in SD. Seed heterogeneity is a well-known phenomenon with several studies addressing its importance. Thus, this article addresses the problems derived from seed heterogeneity by the development of the single-seed RNA-Seq method. (Summary by Eva Maria Gomez Alvarez, @eva_ga96). Plant Physiol. 10.1093/plphys/kiac265
Secondary dormancy (SD) of seeds is a natural strategy that allows survival of the plant in environments with unfavourable climatic conditions. However, in Arabidopsis, a variance of dormancy depth in between identical seeds can be explained by population-based threshold models. This single-seed variability relies on the differences in oxygen consumption, hormone levels and sensitivity, and endo-β-mannanase enzymatic activity. In particular, the single-seed response to SD has been less studied than the response to primary dormancy. In this work, Krzyszton et al., describe a protocol for single-seed transcriptomic analysis to show differences between genetically identical seeds that are subjected to the same stress. The authors subjected Arabidopsis thaliana seeds to high temperature and darkness to induce SD. They observed that the treatment generates the same transcriptomic drop during the stress in all the seedlings. However, transcriptomic differences in the recovery step were observed between the germinating seeds and the ones that remained in SD. Seed heterogeneity is a well-known phenomenon with several studies addressing its importance. Thus, this article addresses the problems derived from seed heterogeneity by the development of the single-seed RNA-Seq method. (Summary by Eva Maria Gomez Alvarez, @eva_ga96). Plant Physiol. 10.1093/plphys/kiac265