Single-plant omics provides transcriptional insights into the transition from the vegetative to reproductive phases
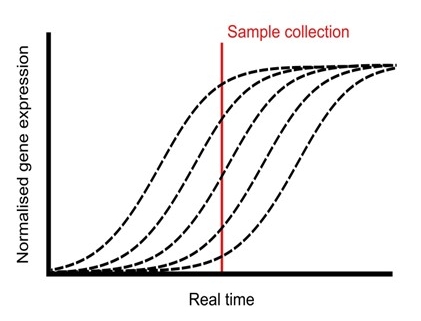
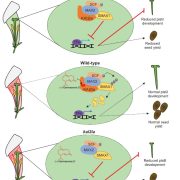
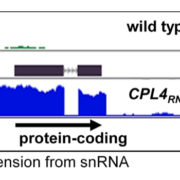
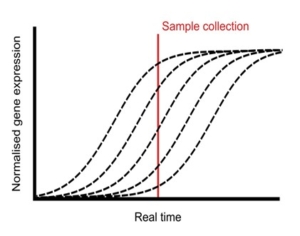 Plants undergo a series of physiological processes when transitioning from the juvenile to the vegetative phase, and then vegetative to the reproductive phase. RNA-Seq offers substantial potential for uncovering the transcriptional landscape underlying these developmental transitions. However, developmental asynchrony among individual plants within a population creates variations in the spatiotemporal expression of genes. Redmond et al. performed a single-plant-omics study on a large population of Arabidopsis thaliana and their investigation revealed the detailed sequence of transcriptional events that occur before and after the bolting transition. Using single-plant-omics allowed the researchers to order individual plants by their intrinsic biological age, providing a high-resolution transcriptional landscape. They trained different models to test whether these models are more or less informative compared to gene expression data. Results revealed that most of the differentially expressed genes were closely linked with the traits of biomass and leaf area. Implication of the pseudotime inference algorithm revealed certain senescence-related traits e.g., transcriptional repression of ribosome biosynthesis followed by photosynthesis shutdown is a major event that serves as a landmark when a bolting plant changes from vegetative to reproductive phase. Their study highlights the role of environmental asynchrony and opens new avenues to use single-plant-omics in synthetic biology and molecular breeding programs. (Summary by Asif Ali @pbgasifkalas) Plant Cell. 10.1093/plcell/koae226
Plants undergo a series of physiological processes when transitioning from the juvenile to the vegetative phase, and then vegetative to the reproductive phase. RNA-Seq offers substantial potential for uncovering the transcriptional landscape underlying these developmental transitions. However, developmental asynchrony among individual plants within a population creates variations in the spatiotemporal expression of genes. Redmond et al. performed a single-plant-omics study on a large population of Arabidopsis thaliana and their investigation revealed the detailed sequence of transcriptional events that occur before and after the bolting transition. Using single-plant-omics allowed the researchers to order individual plants by their intrinsic biological age, providing a high-resolution transcriptional landscape. They trained different models to test whether these models are more or less informative compared to gene expression data. Results revealed that most of the differentially expressed genes were closely linked with the traits of biomass and leaf area. Implication of the pseudotime inference algorithm revealed certain senescence-related traits e.g., transcriptional repression of ribosome biosynthesis followed by photosynthesis shutdown is a major event that serves as a landmark when a bolting plant changes from vegetative to reproductive phase. Their study highlights the role of environmental asynchrony and opens new avenues to use single-plant-omics in synthetic biology and molecular breeding programs. (Summary by Asif Ali @pbgasifkalas) Plant Cell. 10.1093/plcell/koae226