Single-nucleus sequencing reveals transition from C3 to C4 photosynthesis
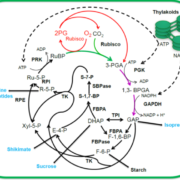
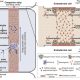
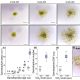
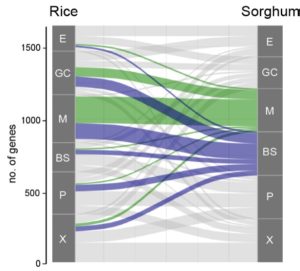 Sometime in the Cretaceous period (dinosaur time!), some monocots acquired a special pathway for carbon fixation, rendering them more efficient particularly in hot or dry environments. In most monocots, carbon is fixed by Rubisco in the mesophyll cells. In the innovative pathway, carbon fixation is split into two cell types, with the initial capture of CO2 taking place in the mesophyll cells by the enzyme PEPC to form a four-carbon product which delivers substrate to Rubisco, localized in the bundle sheath cell; this pathway is called C4 to reflect the novel four-carbon intermediate. To understand the origin of the C4 pathway, Swift and Luginbuehl et al. conducted a transcriptome comparison between rice (C3) and sorghum (C4) plants, with a particular focus on mesophyll and bundle sheath cells. The authors started by RNA sequencing of isolated nuclei, eliminating the problems associated with tissue-level transcriptomics and providing increased granularity. One of the main results of this elegant work is that genes that are normally expressed in mesophyll cells in rice are now active in the bundle sheath cells of sorghum, accomplished by the genes acquiring the relevant transcription factor binding sites (cis-elements). These data not only provide a glimpse into the evolution of C4 photosynthesis, but provide important clues for the efforts to engineer the more efficient C4 pathway into C3 plants such as rice. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.10.26.562893
Sometime in the Cretaceous period (dinosaur time!), some monocots acquired a special pathway for carbon fixation, rendering them more efficient particularly in hot or dry environments. In most monocots, carbon is fixed by Rubisco in the mesophyll cells. In the innovative pathway, carbon fixation is split into two cell types, with the initial capture of CO2 taking place in the mesophyll cells by the enzyme PEPC to form a four-carbon product which delivers substrate to Rubisco, localized in the bundle sheath cell; this pathway is called C4 to reflect the novel four-carbon intermediate. To understand the origin of the C4 pathway, Swift and Luginbuehl et al. conducted a transcriptome comparison between rice (C3) and sorghum (C4) plants, with a particular focus on mesophyll and bundle sheath cells. The authors started by RNA sequencing of isolated nuclei, eliminating the problems associated with tissue-level transcriptomics and providing increased granularity. One of the main results of this elegant work is that genes that are normally expressed in mesophyll cells in rice are now active in the bundle sheath cells of sorghum, accomplished by the genes acquiring the relevant transcription factor binding sites (cis-elements). These data not only provide a glimpse into the evolution of C4 photosynthesis, but provide important clues for the efforts to engineer the more efficient C4 pathway into C3 plants such as rice. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2023.10.26.562893