Single cell transcriptomics aids gene discovery of complex natural product biosynthesis
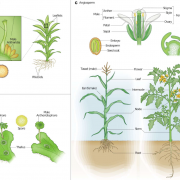
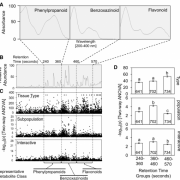
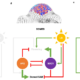
 From an ancient Greek cure-all to a modern treatment for mild depression, Hypericum perforatum (St. John’s wort) is a fascinating weed. Its leaves and flowers produce hyperforin, a metabolite derived from the isoprenoid pathway, which acts as a serotonin reuptake inhibitor. Despite partial genome and transcriptome data, the later steps in its biosynthesis pathway remain unclear. In a recent publication in Molecular Plant, Wu and coauthors aimed to uncover these missing steps. They used whole genome sequencing together with single-cell RNA sequencing (scRNA-seq) of leaf- and flower-derived protoplasts to identify a cell cluster with high expression of known biosynthesis genes, denominated “Hyper cells”. GO analysis revealed four putative genes expressed in these Hyper cells for the elusive last steps of the pathway. These candidates, called HpPT1 to 4, were cloned and reconstituted in yeast, where they were shown to catalyze the different prenylation reactions, which was further validated using a transient expression system in planta. Their study explains previous difficulties in gene discovery, as candidate genes were likely masked using bulk RNA-sequencing. In conclusion, the authors provide a complete elucidation of the hyperforin biosynthesis pathway and successful production of hyperforin in yeast which offers tremendous possibilities for the optimization of hyperforin heterologous cell factories. In addition, this study nicely highlights the power of scRNA-seq in overcoming challenges in gene discovery, apart from its classical applications. (Summary by Thomas Depaepe @thdpaepe). Molecular Plant 10.1016/j.molp.2024.08.003.
From an ancient Greek cure-all to a modern treatment for mild depression, Hypericum perforatum (St. John’s wort) is a fascinating weed. Its leaves and flowers produce hyperforin, a metabolite derived from the isoprenoid pathway, which acts as a serotonin reuptake inhibitor. Despite partial genome and transcriptome data, the later steps in its biosynthesis pathway remain unclear. In a recent publication in Molecular Plant, Wu and coauthors aimed to uncover these missing steps. They used whole genome sequencing together with single-cell RNA sequencing (scRNA-seq) of leaf- and flower-derived protoplasts to identify a cell cluster with high expression of known biosynthesis genes, denominated “Hyper cells”. GO analysis revealed four putative genes expressed in these Hyper cells for the elusive last steps of the pathway. These candidates, called HpPT1 to 4, were cloned and reconstituted in yeast, where they were shown to catalyze the different prenylation reactions, which was further validated using a transient expression system in planta. Their study explains previous difficulties in gene discovery, as candidate genes were likely masked using bulk RNA-sequencing. In conclusion, the authors provide a complete elucidation of the hyperforin biosynthesis pathway and successful production of hyperforin in yeast which offers tremendous possibilities for the optimization of hyperforin heterologous cell factories. In addition, this study nicely highlights the power of scRNA-seq in overcoming challenges in gene discovery, apart from its classical applications. (Summary by Thomas Depaepe @thdpaepe). Molecular Plant 10.1016/j.molp.2024.08.003.