Shooting for the STARRs: A Modified STARR-seq Assay for Rapid Identification and Evaluation of Plant Regulatory Sequences in Tobacco Leaves
A single genome gives rise to different cell types and organs in response to precise temporal and spatial regulation of gene expression, driven by developmental and environmental cues. These expression patterns are orchestrated by cis-regulatory elements, distal enhancers and gene-proximal promoters. Active enhancer elements can be identified by Self-Transcribing Active Regulatory Region Sequencing (STARR-seq), a massively parallel enhancer reporter assay initially developed in Drosophila (Arnold et al., 2014) and previously applied in rice and maize protoplasts (Sun et al., 2019; Ricci et al., 2019).
STARR-seq relies on the insertion of candidate enhancer libraries in the 3’ untranslated region (3’-UTR) of a reporter gene controlled by a minimal promoter. Regulatory activity of the tested sequence translates into self-transcription, quantified by RNA-Seq. STARR-seq has discovered thousands of putative enhancer elements and confirmed known ones in rice and maize genomes. A major limitation of the method resides in its protoplast- and transfection-dependency, hindering its use in plant systems not amenable to such techniques. Due to its inherent disconnection from the plant developmental context, the protoplast STARR-seq system might also be less amenable to identifying development-specific and context-responsive enhancers, both of which are of particular interest for plant breeding and genetic engineering.
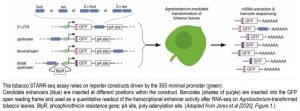 In a new study, Jores et al. bypass these limitations by establishing an optimized STARR-seq assay in tobacco. Transient expression of the STARR-seq libraries is achieved by efficient Agrobacterium-mediated transformation through syringe-infiltration of intact tobacco leaves. To validate the specificity and sensitivity of the STARR-seq assay in tobacco, the authors designed a reporter construct containing a barcoded GFP reporter gene under the control of a Cauliflower mosaic virus 35S minimal promoter and a 35S core enhancer (Figure). Candidate enhancer elements can be inserted at different positions within the construct. Each candidate enhancer is uniquely linked to a barcoded GFP reporter, providing a specific and quantitative readout for the activity of the element revealed by RNA-seq.
In a new study, Jores et al. bypass these limitations by establishing an optimized STARR-seq assay in tobacco. Transient expression of the STARR-seq libraries is achieved by efficient Agrobacterium-mediated transformation through syringe-infiltration of intact tobacco leaves. To validate the specificity and sensitivity of the STARR-seq assay in tobacco, the authors designed a reporter construct containing a barcoded GFP reporter gene under the control of a Cauliflower mosaic virus 35S minimal promoter and a 35S core enhancer (Figure). Candidate enhancer elements can be inserted at different positions within the construct. Each candidate enhancer is uniquely linked to a barcoded GFP reporter, providing a specific and quantitative readout for the activity of the element revealed by RNA-seq.
The authors first tested the dynamic range of the tobacco STARR-seq assay by systematically analyzing the positional effect of the 35S enhancer on reporter gene transcription. As expected, the 35S enhancer is orientation-insensitive and exerts distance-dependent transcriptional enhancement when located either upstream or downstream of the reporter gene. Addition of an extra 35S enhancer copy appreciably increased the transcriptional output, with a stronger effect for downstream and distal upstream enhancers. By contrast, the 35S enhancer displayed minimal-to-null activity when located in transcribed regions such as the 3’-UTR, in contrast with previous reports in rice and maize protoplasts. To address whether this finding was specific to the tobacco system, the authors transformed maize protoplasts, finding again that placement within the transcribed region resulted in little signal.
To validate the sensitivity of the assay in condition-specific contexts, the authors selected three known light-sensitive plant enhancers (AB80, Cab-1, and rbcS-E9) to test their activity using tobacco STARR-Seq. The method accurately detected light-dependent changes in transcriptional activity induced by either intact enhancer sequences or a fragment library derived from a plasmid containing the aforementioned enhancers. Using a fragment library of shorter size (84bp vs. 191bp on average) allowed the identification of multiple potential functional sub-motifs within the tested enhancers, revealing that the tobacco STARR-seq assay can accurately pinpoint functional cis-regulatory elements.
The authors further tested the suitability of the assay for the functional dissection of such sub-domains through saturation mutagenesis. To answer this question, all possible mutation within the 35S promoter and enhancer were generated, and the effect of individual mutations on the reporter gene transcription scored in both contexts. Mutations in the 35S promoter showed a weak-to-no effect on transcription, with the exception of the TATA box. In contrast, saturation mutagenesis of the 35S enhancer identified multiple nucleotides critical for the activity of the cis-regulatory element, thus revealing functional sub-motifs within the enhancer. Most of the functional sub-motifs overlapped with transcription factor binding sites. Lastly, systematic combinations of the functional sub-domains forming synthetic enhancers demonstrated modular and non-additive enhancer activity, supporting the potential of the tobacco STARR-seq assay for building synthetic regulatory elements.
The high conservation of transcription factors in the plant lineage paves the way for the tobacco STARR-seq system to be a powerful and versatile approach for in-depth dissection of regulatory sequences in many plant genomes. The continuous development of such methods opens new possibilities in the genetic engineering of regulatory elements and ultimately in the dissection of the gene regulatory networks orchestrating plant development.
Matthias Benoit
Howard Hughes Medical Institute
Cold Spring Harbor Laboratory
ORCID: 0000-0002-3958-3173
REFERENCES
Arnold, C.D., Gerlach, D., Spies, D., Matts, J.A., Sytnikova, Y.A., Pagani, M., Lau, N.C., and Stark, A. (2014). Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat. Genet. 46: 685–692.
Ricci, W.A. et al. (2019). Widespread long-range cis-regulatory elements in the maize genome. Nat. Plants 5: 1237–1249.
Sun, J., He, N., Niu, L., Huang, Y., Shen, W., Zhang, Y., Li, L., and Hou, C. (2019). Global Quantitative Mapping of Enhancers in Rice by STARR-seq. Genomics. Proteomics Bioinformatics 17: 140–153.
Jores, T., Tonnies, J., Dorrity, M.W., Cuperus, J.T., Fields, S. and Queitsch, C. (2020). Identification of plant enhancers and their constituent elements by STARR-seq in tobacco leaves. The Plant Cell. https://doi.org/10.1105/tpc.20.00155