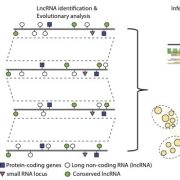
Shoot and root single cell sequencing reveals tissue- and daytime-specific transcriptome profiles (Plant Physiol.)
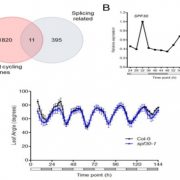
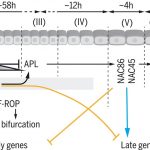
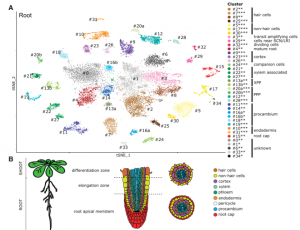 Many biological processes are controlled by the circadian clock in plants, yet studies often don’t report sampling timepoints, leaving much to be discovered about the plant circadian rhythm. Apelt et al. have utilised single cell RNA sequencing to examine almost 70,000 Arabidopsis cells from above and below ground tissues at day-end and night-end timepoints to reveal how the circadian clock influences the plant transcriptome. Transcriptomic changes in subterranean and above ground tissues varied with sampling period, leading to identification of day and night tissue-specific markers. MERCY1, a novel marker gene for dividing cells in both shoot and root tissues, may also influence flowering time, bolting and root development. This work provides the research community with a high-resolution transcriptomic map of single cells from shoot tissue for day and night and root tissue at an end of night timepoint. (Summary by Orla Sherwood @orlasherwood) Plant Physiol. 10.1093/plphys/kiab537
Many biological processes are controlled by the circadian clock in plants, yet studies often don’t report sampling timepoints, leaving much to be discovered about the plant circadian rhythm. Apelt et al. have utilised single cell RNA sequencing to examine almost 70,000 Arabidopsis cells from above and below ground tissues at day-end and night-end timepoints to reveal how the circadian clock influences the plant transcriptome. Transcriptomic changes in subterranean and above ground tissues varied with sampling period, leading to identification of day and night tissue-specific markers. MERCY1, a novel marker gene for dividing cells in both shoot and root tissues, may also influence flowering time, bolting and root development. This work provides the research community with a high-resolution transcriptomic map of single cells from shoot tissue for day and night and root tissue at an end of night timepoint. (Summary by Orla Sherwood @orlasherwood) Plant Physiol. 10.1093/plphys/kiab537