Shared Genetic Control of Root Traits across Taxa
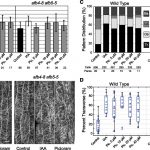
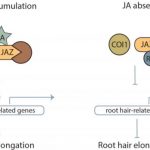
Root system architecture (RSA) plays a crucial role in plant productivity and tolerance to environmental stresses. The maize (Zea mays) root 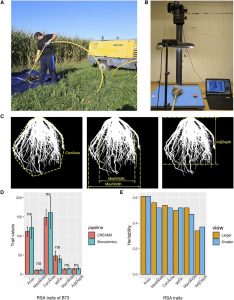 system, composed of the embryonic primary root and variable numbers of seminal roots, as well as postembryonic shoot-borne and lateral roots, is both different and more complex than the model plant Arabidopsis. [The sorghum (Sorghum bicolor) root system is similar to maize although it lacks seminal roots.] Based on the fundamental morphological differences between grasses and Arabidopsis, it is unlikely that a complete understanding of the genetic regulation of RSA in grasses can be elucidated from studies of Arabidopsis. Genome-wide associated study (GWAS) offers the opportunity to identify genes affecting natural variation of quantitative traits via the association of markers across the genome with phenotypic variation within diversity panels. With the ready availability of large numbers of genetic markers, phenotyping has become the bottleneck for GWAS. To overcome this bottleneck, Zheng et al. (10.1104/pp.19.00752) have developed a high-throughput pipeline for the cleaning of field-grown roots, as well as a semiautomated pipeline for the extraction of RSA traits from images. Comparative analyses of maize and sorghum GWAS results provide strong evidence for shared genetic control of RSA in these two species and the conservation of functional roles for syntenic orthologous gene pairs.
system, composed of the embryonic primary root and variable numbers of seminal roots, as well as postembryonic shoot-borne and lateral roots, is both different and more complex than the model plant Arabidopsis. [The sorghum (Sorghum bicolor) root system is similar to maize although it lacks seminal roots.] Based on the fundamental morphological differences between grasses and Arabidopsis, it is unlikely that a complete understanding of the genetic regulation of RSA in grasses can be elucidated from studies of Arabidopsis. Genome-wide associated study (GWAS) offers the opportunity to identify genes affecting natural variation of quantitative traits via the association of markers across the genome with phenotypic variation within diversity panels. With the ready availability of large numbers of genetic markers, phenotyping has become the bottleneck for GWAS. To overcome this bottleneck, Zheng et al. (10.1104/pp.19.00752) have developed a high-throughput pipeline for the cleaning of field-grown roots, as well as a semiautomated pipeline for the extraction of RSA traits from images. Comparative analyses of maize and sorghum GWAS results provide strong evidence for shared genetic control of RSA in these two species and the conservation of functional roles for syntenic orthologous gene pairs.