Shaping pathogenicity: How CRISPR-Cas loss fuels Xanthomonas evolution
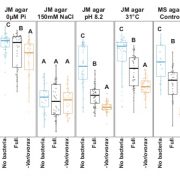
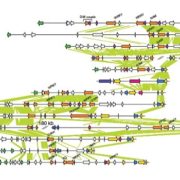
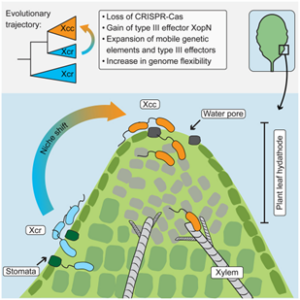 Pathogens have evolved diverse infection strategies governed by virulence factors, often targeting specific host organs or tissues. Genome fluidity plays a crucial role in enabling microbial pathogens to adapt to the dynamic selection pressures imposed by co-evolution with their hosts. In a recent study, Paauw and colleagues used a pangenome approach to investigate the genetic factors and evolutionary processes shaping the evolution of pathogenic Xanthomonas campestris (Xc) in a collection of over 180 isolates from diseased Brassica plants. Notably, different Xc pathovars exhibit distinct tissue specificities within the same leaf: Xc pv. campestris (Xcc) infects the vasculature, whereas Xc pv. raphanin (Xcr) enters leaves exclusively through stomata, like most bacteria. However, the genetic mechanisms driving the diversification of virulence factors remain unclear. Prokaryotic CRISPR-Cas systems serve as adaptive immune defenses that restrict horizontal gene transfer and limit the uptake of foreign DNA. The authors found that loss of CRISPR-Cas is more prominent in the Xcc genome compared to Xcr, facilitating the acquisition of virulence factors, and increasing genome plasticity. This enhanced genomic flexibility allows Xcc to colonize additional plant tissues that other pathovars cannot. While gene regulations can occur rapidly during host infection, the emergence of new species or subspecies requires continuous genome evolution. Overall, these findings suggest that CRISPR-Cas loss has been a key driver of Xc evolution, contributing to the emergence of agronomically important vascular pathogens in cabbage. (Summary by Ching Chan @ntnuchanlab) Curr. Biol. 10.1016/j.cub.2025.01.003
Pathogens have evolved diverse infection strategies governed by virulence factors, often targeting specific host organs or tissues. Genome fluidity plays a crucial role in enabling microbial pathogens to adapt to the dynamic selection pressures imposed by co-evolution with their hosts. In a recent study, Paauw and colleagues used a pangenome approach to investigate the genetic factors and evolutionary processes shaping the evolution of pathogenic Xanthomonas campestris (Xc) in a collection of over 180 isolates from diseased Brassica plants. Notably, different Xc pathovars exhibit distinct tissue specificities within the same leaf: Xc pv. campestris (Xcc) infects the vasculature, whereas Xc pv. raphanin (Xcr) enters leaves exclusively through stomata, like most bacteria. However, the genetic mechanisms driving the diversification of virulence factors remain unclear. Prokaryotic CRISPR-Cas systems serve as adaptive immune defenses that restrict horizontal gene transfer and limit the uptake of foreign DNA. The authors found that loss of CRISPR-Cas is more prominent in the Xcc genome compared to Xcr, facilitating the acquisition of virulence factors, and increasing genome plasticity. This enhanced genomic flexibility allows Xcc to colonize additional plant tissues that other pathovars cannot. While gene regulations can occur rapidly during host infection, the emergence of new species or subspecies requires continuous genome evolution. Overall, these findings suggest that CRISPR-Cas loss has been a key driver of Xc evolution, contributing to the emergence of agronomically important vascular pathogens in cabbage. (Summary by Ching Chan @ntnuchanlab) Curr. Biol. 10.1016/j.cub.2025.01.003