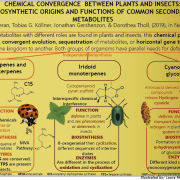
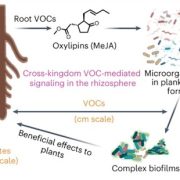
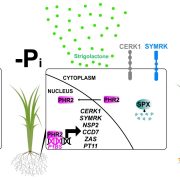
Arabidopsis thaliana-associated Pseudomonas diversity and evolution
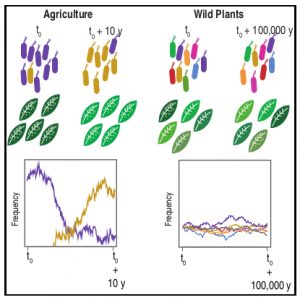 There is not much available information regarding the evolutionary aspects of one of the most studied pathosystem in plant biology: Arabidopsis thaliana and Pseudomonas. Karazov et al. performed various surveys in Arabidopsis wild populations, for the detection and characterization of their associated Pseudomonas strains. Pseudomonas spp. were detected in 97% of epi- and 88% of endophytic samples, and classified into 56 operational taxonomic units (OTUs) according to 16S sequence similarity. OTU5, classified as P. viridiflava, was found in 59% of epi- and 58% of endophytic samples, making it the predominant lineage within the isolates. Whole genome sequence of the OTU5 isolates led to the classification of 1355 isolates into 82 different strains, closely related to P. viridiflava and P. syringae. Five from the OTU5 strains accounted for 46% of the sequenced isolates, while non-OTU5 strains were rarely overrepresented in several isolates. 26 of the OTU5 strains were tested for their pathogenicity in the Eyach 15-2 Arabidopsis genotype; all but one of the assessed strains reduced plant growth, and some of these strains were found to be even more virulent than the highly virulent Arabidopsis pathogen P. syringae pv. tomato DC3000. Finally, through phylogenetic analyses, the authors were able to predict that strains belonging to OTU5 diverged approximately 300,000 years ago, maintaining nonetheless their ability to successfully colonize Arabidopsis, and contributing to a high microbial diversity in wild populations of this plant. (Summary by Juan S. Ramirez-Prado) Cell Host Microb. 10.1016/j.chom.2018.06.011
There is not much available information regarding the evolutionary aspects of one of the most studied pathosystem in plant biology: Arabidopsis thaliana and Pseudomonas. Karazov et al. performed various surveys in Arabidopsis wild populations, for the detection and characterization of their associated Pseudomonas strains. Pseudomonas spp. were detected in 97% of epi- and 88% of endophytic samples, and classified into 56 operational taxonomic units (OTUs) according to 16S sequence similarity. OTU5, classified as P. viridiflava, was found in 59% of epi- and 58% of endophytic samples, making it the predominant lineage within the isolates. Whole genome sequence of the OTU5 isolates led to the classification of 1355 isolates into 82 different strains, closely related to P. viridiflava and P. syringae. Five from the OTU5 strains accounted for 46% of the sequenced isolates, while non-OTU5 strains were rarely overrepresented in several isolates. 26 of the OTU5 strains were tested for their pathogenicity in the Eyach 15-2 Arabidopsis genotype; all but one of the assessed strains reduced plant growth, and some of these strains were found to be even more virulent than the highly virulent Arabidopsis pathogen P. syringae pv. tomato DC3000. Finally, through phylogenetic analyses, the authors were able to predict that strains belonging to OTU5 diverged approximately 300,000 years ago, maintaining nonetheless their ability to successfully colonize Arabidopsis, and contributing to a high microbial diversity in wild populations of this plant. (Summary by Juan S. Ramirez-Prado) Cell Host Microb. 10.1016/j.chom.2018.06.011