Semi-annotated metabolomics facilitates specialized metabolic gene and pathway elucidation
Anting Zhu et al. establish an approach to identify metabolites and their corresponding genes and pathways in wheat.
https://doi.org/10.1093/plcell/koad286
By A. Zhu, M. Liu, Z. Tian, W. Liu, W. Chen et al. National Key Laboratory of Crop Genetic Improvement and National Center of Plant Gene Research, Huazhong Agricultural University.
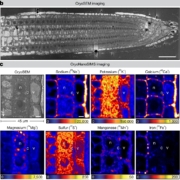
Background: Plants produce many metabolites that regulate their biological processes. Fully dissecting the structures of these metabolites is complex and time-consuming and is also a bottleneck in the applications of metabolomics. Structurally, metabolites are often “variations on a theme” resulting from one or more modifications to a common backbone structure. These modification groups are relatively simple, small, limited in quantity, and easy to annotate. Most importantly, they are catalyzed by tailoring enzymes in plants. Bread wheat is a staple food crop consumed worldwide; its large genome hinders the identification of gene resources for crop improvement.
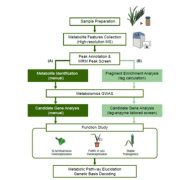
Question: Can the relationship between metabolite modification structures and enzyme reactions be incorporated into metabolite-genome wide association studies to identify metabolites and their corresponding genes and pathways in wheat?
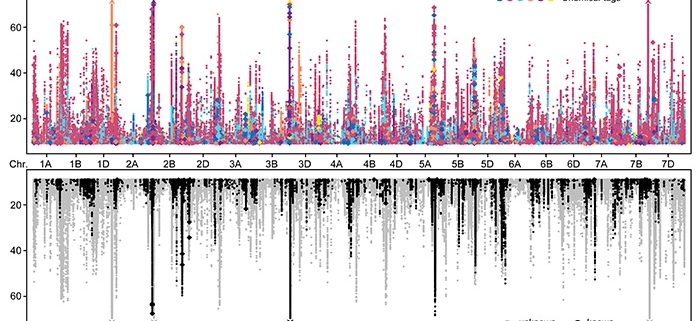
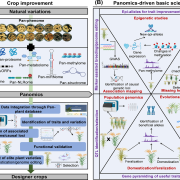
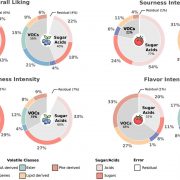
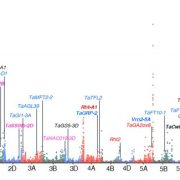
F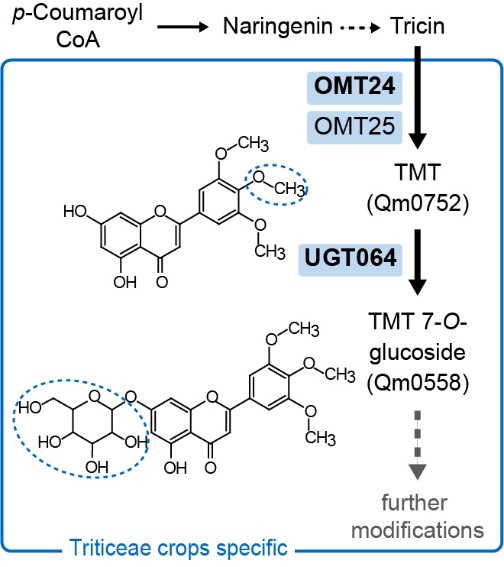 indings: We profiled metabolic fragment enrichment features in wheat leaves and chose six common modifications as tags to apply a semi-annotated metabolic approach in a genome-wide association study (mGWAS) in wheat. Compared to the traditional mGWAS workflow, our strategy is more efficient, unbiased, and outstanding at discovering species-specific metabolites and their corresponding genes and pathways. Through our approach, we obtained a candidate list that offered vast gene resources for follow-up studies. From the list, we investigated a gene cluster that encodes a Triticeae crop-specific flavonoid pathway that probably plays an important role in stress resistance.
indings: We profiled metabolic fragment enrichment features in wheat leaves and chose six common modifications as tags to apply a semi-annotated metabolic approach in a genome-wide association study (mGWAS) in wheat. Compared to the traditional mGWAS workflow, our strategy is more efficient, unbiased, and outstanding at discovering species-specific metabolites and their corresponding genes and pathways. Through our approach, we obtained a candidate list that offered vast gene resources for follow-up studies. From the list, we investigated a gene cluster that encodes a Triticeae crop-specific flavonoid pathway that probably plays an important role in stress resistance.
Next steps: We are working on the applications of this approach to other crops, verifying more candidates, constructing related pathways, and identifying new metabolites for the ultimate goal of crop improvement.
Reference:
Anting Zhu, Mengmeng Liu, Zhitao Tian, Wei Liu, Xin Hu, Min Ao, Jingqi Jia, Taotao Shi, Hongbo Liu, Dongqin Li, Hailiang Mao, Handong Su, Wenhao Yan, Qiang Li, Caixia Lan, Alisdair R. Fernie and Wei Chen (2023). Chemical-tag-based semi-annotated metabolomics facilitates gene identification and specialized metabolic pathway elucidation in wheat. https://doi.org/10.1093/plcell/koad286